This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Reducing contamination in single-molecule DNA extraction using nanopore technology
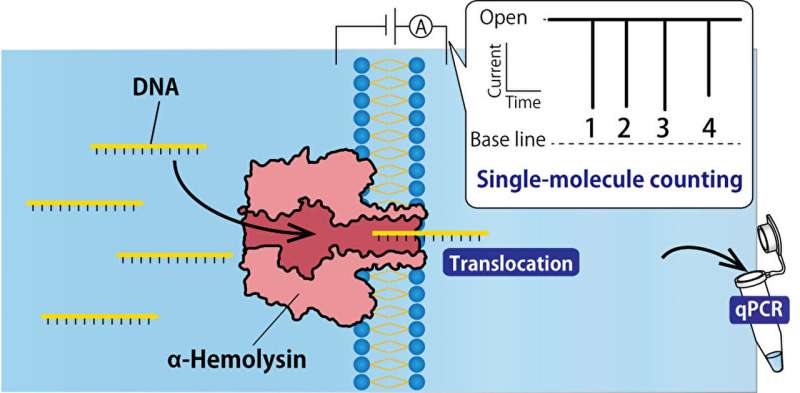
Single-molecule DNA detection using nanopore technology offers real-time analysis of DNA and RNA strands. It is a low-cost and flexible technique that can be used in clinical and research settings when samples need to be analyzed quickly and efficiently. However, this is new technology that still has some drawbacks, such as sample contamination.
Research to improve and optimize single-molecule DNA extraction with nanopores is ongoing. A recently published paper outlines how a DNA-filtering system using a nanopore developed from the α-hemolysin protein.
The paper was published on June 6 in Analytical Chemistry.
"We set out to develop a DNA filtering system using α-hemolysin (αHL) nanopores and to better understand the challenges faced in achieving accurate single-molecule counting," said Ryuji Kawano, a professor at Tokyo University of Agriculture and Technology in Tokyo, Japan. "The development of a DNA filtering system using αHL nanopores has the potential to revolutionize DNA analysis by enabling real-time detection and analysis of DNA at the single-molecule level, eliminating the need for labeling or partitioning sample solutions."
In this technique, a microdevice is used to prepare two water-in-oil droplets. One is the sample droplet containing target DNAs and one is the controlling droplet without the DNAs. The droplets are separated by a lipid bilayer, which allows a single molecule of DNA to move through an αHL nanopore. As the molecules move through this nanopore, they are counted as electrical signals. Researchers looked at ways this technique could be improved.
"We aimed to optimize the experimental environment, reduce the volume of solution containing the target molecule, and use the PCR clamp method to tackle the issue of contamination, which we found to be an almost insolvable problem in single-molecule counting," said Kawano.
To solve the problem of contamination, researchers proposed several different avenues. Contamination in DNA samples can come from the air and from impurities in the samples. Improving air-borne contamination proved to be relatively straightforward.
The sample DNA was used in one space that was designated as an experimental area, special pipettes that limited airborne contamination were used, and work areas were decontaminated with UV light for 15 minutes. DNA contamination from the air was completely prevented after implementing these changes.
However, DNA contamination also happens within the oil and lipid mixture and solving the problem of this contamination was less straightforward. Researchers tried different phospholipids, but this did not have an impact on contamination. Another technique called the contact bubble bilayer method, in which water bubbles are introduced to the oil layer using glass pipettes, was tried, but contamination was detected. This is likely due to DNA molecules being accidentally introduced to the oil and lipid mixture when the emulsion is formed.
Finally, researchers tested the PCR clamp method, which adds peptide nucleic acids (PNA) to bind to the DNA, creating PNA-DNA duplexes. As the duplex moves through the nanopore, they unzip and only the target DNA moves to the next droplet. The same method is applied to contaminant DNA. PNA is added to the solution so it can bind to the contaminants, but this PNA-DNA duplex is not unzipped. This makes it easy to tell the target DNA from the contaminant DNA, allowing the results to exclude and disregard the extra DNA.
When PNAs were added to the solution, there was a 99.98% reduction in DNA contamination.
Looking ahead, researchers hope to continue optimizing and refining the DNA filtering system. "We aim to overcome the challenges associated with accurate single-molecule counting, particularly the issue of contamination. Our ultimate goal is to develop a reliable and efficient single-molecule filter," said Kawano.
More information: Asuka Tada et al, Nanopore Filter: A Method for Counting and Extracting Single DNA Molecules Using a Biological Nanopore, Analytical Chemistry (2023). DOI: 10.1021/acs.analchem.3c00573
Journal information: Analytical Chemistry
Provided by Tokyo University of Agriculture and Technology