This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists develop new AI tool for gene discovery in clinical and research settings
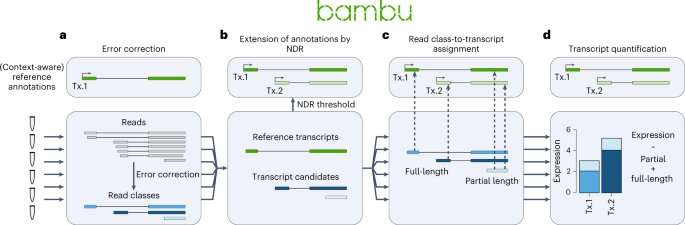
Scientists from A*STAR's Genome Institute of Singapore (GIS) have developed a new tool, named Bambu, which uses artificial intelligence to identify and characterize new genes, enabling an adaptable analysis across various species and samples. With a better understanding of which and how genes are expressed in samples, Bambu provides a better understanding of how cells function.
It is a long-read RNA sequencing tool that can be used in both clinical and research settings to discover how DNA encodes novel transcripts and quantifies them. This innovative tool is named after the bamboo plant, which has extremely long reeds that are analogous to the long reads that Bambu uses. A study detailing the methodology and evaluation of Bambu was published in Nature Methods.
The human genome, which comprises 3.2 billion letters, also known as base pairs, is dwarfed by the lungfish genome with 43 billion, and even more so by the Japanese flower Pari japonica with 149 billion base pairs. Despite a human's relatively smaller genome, there are over 140,000 unique ways genes are encoded within—also referred to as a gene's transcripts—and given the complexity of the body's organs, life stages and responses to perturbations such as diseases, it is estimated that there are many yet to be identified.
This is not only limited to humans, as scientists have been researching organisms such as the durian and Singapore's national flower—and there remains a whole frontier of new genes to be discovered.
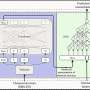
In order to explore the unknown parts of genomes, be it for human, fish or flowers, A*STAR's researchers developed Bambu, which uses long-read RNA sequencing to identify and quantify transcripts.
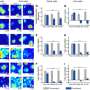
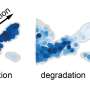
Bambu employs a machine-learning model to rank the likelihood of candidate transcripts representing biologically relevant products. It can identify new transcripts and quantify them with a high degree of precision and sensitivity, providing a more comprehensive understanding of an organism's genetic makeup.
This will allow researchers to identify new roleplayers, such as genes, proteins, and other elements in their field of research and expand their ability to research organisms that are currently under-studied. Furthermore, the discovery of new genes, especially from clinical samples, can lead to the identification of biomarkers for the early detection of diseases or as targets of therapeutics.
An early release of Bambu has been benchmarked by two independent pre-print studies where it is shown to be a top performer among its contemporaries.
"It is fascinating to see that scientists are still discovering new genes even in genomes that have been studied for many years, such as the human or mouse genome. However, the key question is if these transcripts are relevant, or they could be artifacts. To address this, Bambu quantifies the probability that a transcript is real, making transcript and gene discovery much more reliable," said Dr. Jonathan Goke, Group Leader of the Laboratory of Computational Transcriptomics at A*STAR's GIS and the corresponding author of the study. He went on to add: "By providing such a measure of confidence, Bambu can more reliably be applied to find new genes that play a role in human diseases such as cancer."
Dr. Andre Sim, Research Fellow at A*STAR's GIS and co-first author of the study remarked, "Identifying new transcript models require numerous decisions. Bambu simplifies this process using its machine learning model, making this task more accessible to the scientific community."
Prof Patrick Tan, Executive Director of A*STAR's GIS, commented, "Annotating genomes is often the first step in modern genetics towards understanding an organism, and as scientists start looking to research new and exciting species, having accurate transcript discovery provided by tools such as Bambu will be essential."
More information: Ying Chen et al, Context-aware transcript quantification from long-read RNA-seq data with Bambu, Nature Methods (2023). DOI: 10.1038/s41592-023-01908-w
Journal information: Nature Methods