This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Important enzyme for the composition of the gut microbiome discovered
The intestinal microbiome, i.e., the community of bacteria and other microorganisms that live in the human gut, has been shown to affect the metabolism and the immune system. We still do not fully understand how the symbiosis between a healthy microbiome and the host is regulated and how bacterial overgrowth with pathogenic germs can be prevented.
A research team led by Professor Christoph Becker-Pauly from the Institute of Biochemistry at Kiel University (CAU) in collaboration with other working groups has now discovered an important mechanism influencing the intestinal microbiome in mouse models. The results were recently published in the journal Science Advances.
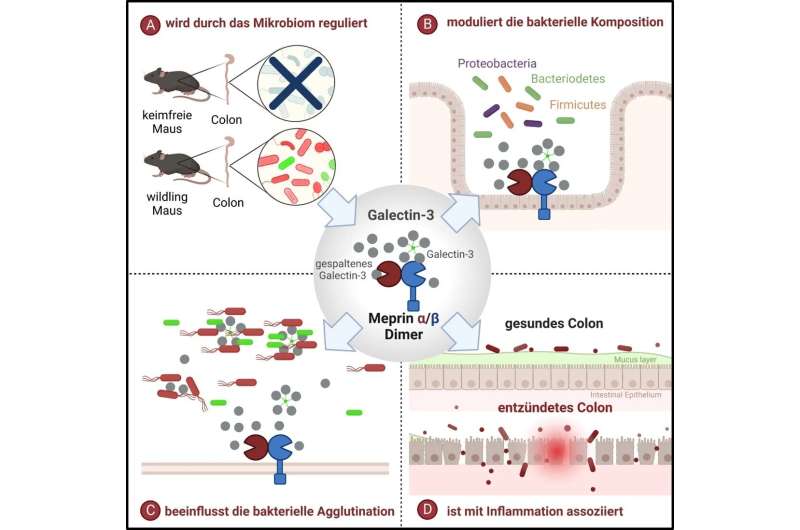
"For the first time, we were able to show that a complex of the enzymes meprin α and meprin β located on intestinal cells influences the microbiome composition by processing the substrate galectin-3," explained first author Cynthia Bülck, Ph.D. student at the CAU's Institute of Biochemistry.
"The activity of this enzyme complex is in turn directly influenced by the microbiome," added Professor Christoph Becker-Pauly, member of the Cluster of Excellence "Precision Medicine in Chronic Inflammation" (PMI) and head of the Unit for Degradomics of the Protease Web. "This means that the microbiome influences host proteins, which in turn influence the microbiome."
The researchers studied this interplay of regulation of the enzyme complex in mouse models with different bacterial colonizations.
Enzymatic cleavage influences binding to bacteria
The research focused on the protein-cleaving enzymes meprin α and meprin β, which are strongly expressed in healthy intestine and are significantly downregulated in chronic inflammatory bowel disease (IBD). "On the one hand, we wanted to elucidate the function of meprins in the small and large intestine, and on the other hand, we wanted to understand how the composition of bacteria in the intestine is fundamentally regulated," said Bülck.
Meprin proteases are found throughout the entire intestine, but they are not typical digestive enzymes. "In previous studies, we were already able to show that these enzymes are responsible for constant detachment and renewing of the mucus layer that protects the small intestinal epithelium," said Becker-Pauly.
In order to clarify the further functions of meprins, which exist as the meprin α/β complex in the large intestine, the researchers first employed a mass spectrometry-based method to search for substrates that are processed by this enzyme complex. "We identified galectin-3 as an important substrate in the large intestine," stated Becker-Pauly.
Galectin-3 is permanently produced in the intestinal villi. It is found both within the cells but also outside the cell in the mucus layer and can interact with bacteria, for example by agglutination. Proteolytic cleavage of galectin-3 by meprin α/β resulted in altered bacterial binding properties. At the same time, depending on the bacterial composition, enzymatic processing of galectin-3 changes. "This means that depending on how many and which bacteria are present, it also has an influence on how much galectin-3 is enzymatically processed," explained Bülck. "The host responds to the microbiome via the cleavage of galectin-3, which is then modulated differently."
The work also showed that the enzymatic cleavage of galectin-3 leads to strong agglutination (clumping) and elimination of the pathogen Pseudomonas aeruginosa, a germ that is responsible for about 10% of all hospital infections in Germany.
Key role in microbiome homeostasis
"Understanding the physiological role of this enzyme complex in the intestine could provide new insights into the development of diseases as well as new ways to prevent and treat intestinal diseases," emphasized Becker-Pauly. In chronic inflammatory bowel diseases, not only the enzyme complex is reduced, but also the substrate galectin-3 is also downregulated. "This may lead to an imbalance of the the intestinal microbiome so that pathogens can spread more easily," suspects the Kiel biochemist. This constellation is certainly not decisive for the development of chronic inflammatory bowel diseases, as many factors play a role in this regard, but it should be considered.
Further research will now focus on elucidating the mechanisms in more detail that lead to the agglutination of certain types of bacteria. In addition, further immunomodulatory substrates of the meprin α/β complex have been identified and their function will now be analyzed.
More information: Cynthia Bülck et al, Proteolytic processing of galectin-3 by meprin metalloproteases is crucial for host-microbiome homeostasis, Science Advances (2023). DOI: 10.1126/sciadv.adf4055
Journal information: Science Advances
Provided by Kiel University