This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
A fast and efficient method for detecting microRNAs
In the early 1990s, scientists who were studying the development of a roundworm identified a small RNA molecule that regulated the expression of specific genes. This marked the discovery of microRNAs (miRNAs), which are now known to be present across all forms of life. As it turns out, these molecules play essential roles in many biological processes.
A few years later, researchers realized that diseases could dysregulate the expression of miRNAs, highlighting their potential as biomarkers. In fact, abnormal miRNA expression is a hallmark of all tumor-related diseases. Thus, miRNA detection techniques may be useful for the early detection of cancer.
However, miRNAs are small and degrade easily, which makes their rapid detection and quantification difficult. To detect miRNAs in a sample, it is usually necessary to first 'amplify' them. Put simply, this means replicating a target miRNA multiple times via biochemical processes so that said miRNA is easier to detect through inexpensive methods. Unfortunately, most state-of-the-art techniques for miRNA amplification can take over five hours to complete, limiting their use in point-of-care testing.
Against this backdrop, a research team, including Associate Professor Chong Zhang from Tsinghua University, China, recently pioneered a new methodology for fast miRNA amplification and detection. As explained in their latest paper, which was published in BioDesign Research, the team combined two well-studied biochemistry techniques into one in a way that greatly reduced the overall time required.
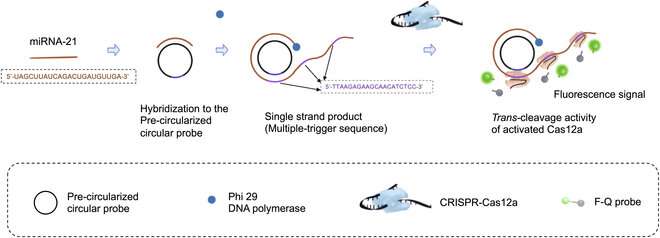
The first technique they used is called rolling circle amplification (RCA). In RCA, the idea is to design a circular DNA molecule or 'probe' to which the target RNA fragment binds. Then, once DNA polymerase enzymes and the necessary DNA building blocks are introduced, the RNA fragment is extended by adding nucleotides complementary to the circular probe. This process results in a long, single strand of genetic material that contains multiple copies of the circular probe.
This is where the second technique, CRISPR-Cas12a, comes into play. The CRISPR-Cas12a is a widely used genetic tool in which a molecular complex is engineered to bind to a specific DNA sequence. In this case, the researchers designed the complex so that it would bind to a region in the complementary sequence to the circular probe.
That is, the CRISPR-Cas12a complexes bound multiple times along the single-strand of DNA that was produced via RCA. Once these complexes were bound, the Cas12a portion activated, splitting a fluorescent probe from its quencher. In turn, this created an easily detectable fluorescent signal that was brighter the more the initial target RNA was amplified.
Besides the combination of these techniques, the researchers improved the reaction time of the RCA step by using 'precircularized probes.' That is, unlike most standard RCA procedures, the probes were given their circular shape prior to the reaction.
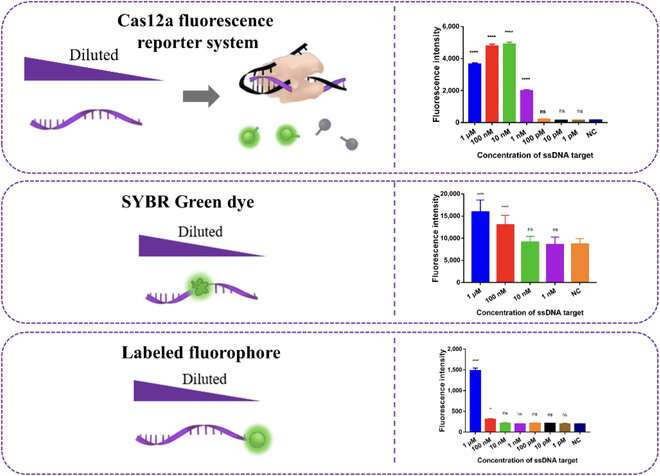
As Prof. Zhang remarks, this made the detection process much quicker without compromising the system's performance: "The detection of miRNA could be completed in only 70 min, rather than the usual five hours, with an excellent limit of detection of 8.1 pM and very high specificity."
Overall, the proposed approach paints a bright future for miRNA detection and their use as biomarkers. Prof. Zhang concludes, "Our design improves the efficiency of CRISPR–Cas and RCA-based sensing strategies and shows great potential in lab-based detection and point-of-care testing." Since the techniques used in this methodology are not prohibitively expensive nor complex to perform, the widespread adoption of the proposed approach in clinical settings is feasible.
More information: Chenqi Niu et al, Exploring the Trans-Cleavage Activity with Rolling Circle Amplification for Fast Detection of miRNA, BioDesign Research (2023). DOI: 10.34133/bdr.0010
Provided by NanJing Agricultural University