This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Trapping and killing superbugs with novel peptide 'nanonets'
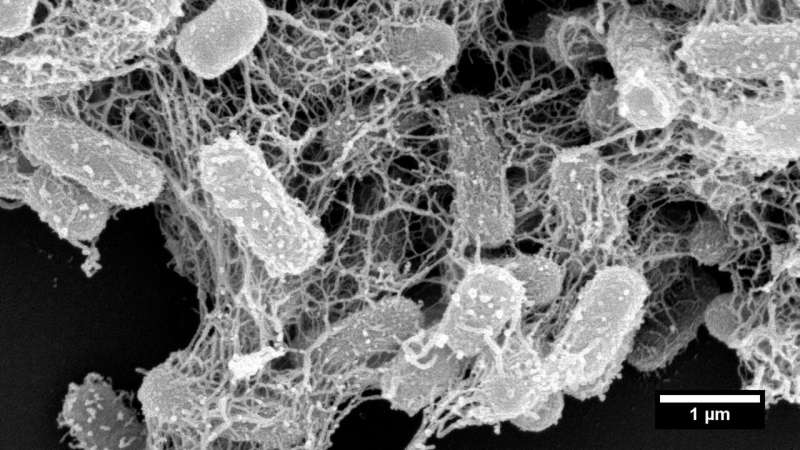
National University of Singapore (NUS) pharmaceutical scientists have developed synthetic peptide nanonets for treating infections by bacteria strains resistant to last-resort antibiotics.
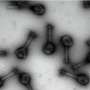
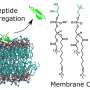
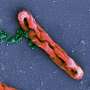
In nature, trap-and-kill is a common immune defense mechanism employed by various species, including humans. In response to the presence of pathogens, peptides are released from host cells and they promptly self-assemble in solution to form cross-linked nanonets, which then entrap the bacteria and render them more vulnerable to antimicrobial components.
Several research groups have explored synthetic biomimetics of nanonets as an avenue for addressing the global healthcare challenge of widespread antibiotic resistance. However, most prominent studies in the field only yielded disjointed short nanofibrils restricted to the bacterial surfaces and are incapable of physically immobilizing the bacteria. Additionally, these designs were lacking in control over the initiation of the self-assembly process.
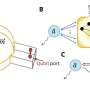
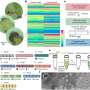
A research team led by Associate Professor Rachel EE from the Department of Pharmacy, NUS has designed short β-hairpin peptides of 15 to 16 residues that are capable of self-assembling into nanonets selectively in response to lipopolysaccharide or lipoteichoic acid, which are integral membrane components unique to bacteria.
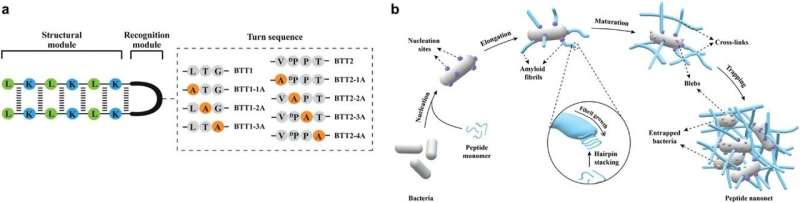
This specificity towards bacteria is an appealing attribute not yet achieved in the field. The peptide nanonets displayed both trapping and antimicrobial killing functionalities, thus offering a direct upgrade from the trap-only nanonets in nature as well as synthetic designs reported in the field. This opens up opportunities for modulating the activity spectrum of the material.
The team further demonstrated functional tunability of the peptides, where potency and fibrillation capacity could be modulated by changing only one or two amino acids at the hairpin turn region of the sequences. Of interest, the nanonets displayed robustness against enzymatic degradation by trypsin, which is a major challenge limiting clinical applications of simple antimicrobial peptides.
Biological evaluations of the peptide nanonets using murine models showed significant antimicrobial efficacy against colistin-resistant bacteria and no systemic toxicity. This work was performed in collaboration with Associate Professor Rajamani Lakshminarayanan who holds joint appointments with the Department of Pharmacy, NUS and the Singapore Eye Research Institute. These findings were published in Advanced Functional Materials.
Prof Ee said, "Our peptide-based nanonets have shown potentials as an alternative anti-infective strategy to address antibiotic resistance. Our next challenge is to optimize the design for clinical application in humans."
In an ongoing effort to fully elucidate the functions of the peptide nanonets, the team is investigating their potential in simultaneously subduing inflammatory responses, a common co-occurrence at the site of bacterial infection.
More information: Nhan Dai Thien Tram et al, Bacteria‐Responsive Self‐Assembly of Antimicrobial Peptide Nanonets for Trap‐and‐Kill of Antibiotic‐Resistant Strains, Advanced Functional Materials (2022). DOI: 10.1002/adfm.202210858
Journal information: Advanced Functional Materials
Provided by National University of Singapore