March 13, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Hidden metabolite–protein interactions captured by MIDAS
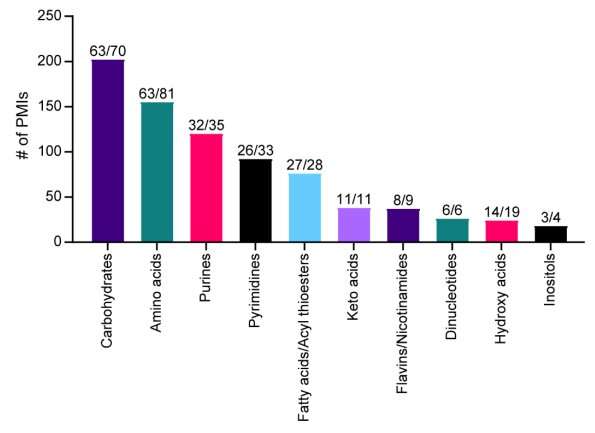
A research group of 46, led by the University of Utah School of Medicine, reports an innovative new method to analyze 33 enzymes from human carbohydrate metabolism and identify 830 protein-metabolite interactions, including known regulators, substrates, and products, as well as previously unreported interactions.
The researchers documented their findings and the new method they used to investigate metabolite–protein interactomes in a paper published in the journal Science.
Within every living cell, proteins and small molecules (metabolites) work together to keep the cell functioning. The metabolite-protein interactome refers to the complex interaction network between metabolites and proteins.
This interactome regulates many vital processes and maintains cellular homeostasis, yet remains an understudied aspect of biology. Recent technological advancements are gradually uncovering the complexity of the protein-metabolite interactome, though it is often through observing general ratios and concentrations of proteins to metabolites, not direct interaction.
One reason direct interactions are challenging to detect is that protein-metabolite interactions are frequently low affinity—they don't remain stuck together well enough to make it through a laboratory process meant to reveal them. So researchers made a better process—mass spectrometry integrated with equilibrium dialysis for the discovery of allostery systematically (MIDAS). Then they put MIDAS to work, looking for metabolite-protein interactions.
Researchers employed a library of 401 compounds representing a sizable fraction of the water-soluble, chemically stable and commercially available components of the human metabolome. By measuring quantity changes between the start and end of the experiment, the researchers could determine the protein metabolite interactions and the binding affinity of those interactions.
The authors write, "A purified protein is separated from a defined library of metabolites by a semipermeable dialysis membrane that allows diffusion of metabolites but not protein. After incubation, the system achieves relative equilibrium, such that the concentration of free metabolites is similar in the protein and metabolite chambers.
"However, the total concentration of those metabolites that interact with the protein is higher or lower in the protein chamber relative to the metabolite chamber dependent on binding affinity and mode of interaction. The protein is then denatured and removed from the protein chamber, and the relative abundances of all metabolites from both chambers is quantified by high-throughput flow injection analysis–mass spectrometry."
A pilot was conducted for validation, using proteins with well-characterized metabolite interactors. In the pilot study, known metabolite interactors were the most frequently detected, validating the method. Three previously undocumented binding events were also discovered, with interesting implications. It may have inadvertently uncovered a feedback regulator of polyamine synthesis, which occurs in some cancers—not a bad start when a pilot validation study makes a mechanistic observation of interest to cancer research.
The team then deployed MIDAS to carbohydrate metabolism enzymes and confirmed previously known substrates, products and regulator interactions. MIDAS also uncovered many previously hidden interactions from diverse metabolic pathways. Therefore, according to the researchers, "MIDAS serves as a conduit to identify, understand, and exploit previously unknown modes of metabolic regulation across the protein-metabolite interactome."
More information: Kevin G. Hicks et al, Protein-metabolite interactomics of carbohydrate metabolism reveals regulation of lactate dehydrogenase, Science (2023). DOI: 10.1126/science.abm3452. www.science.org/doi/10.1126/science.abm3452
Journal information: Science
© 2023 Science X Network