This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Biomolecular sliding at the nanoscale
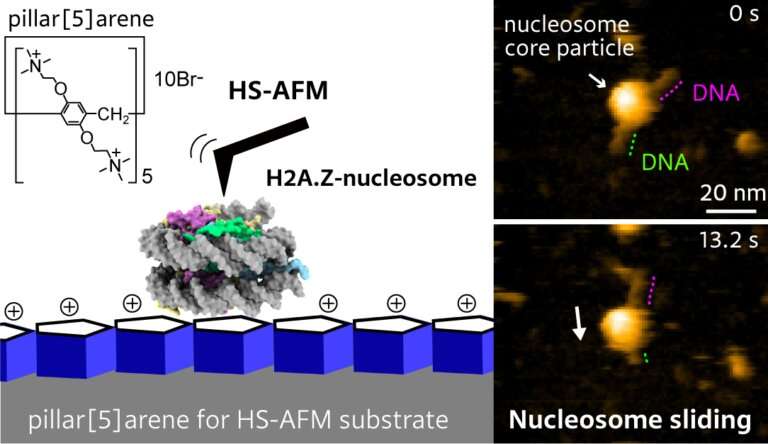
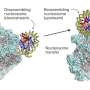
In organisms whose cells have a nucleus, like plants and animals, the basic packaging units of DNA are the so-called nucleosomes. A nucleosome consists of a segment of DNA wound around eight proteins known as histones. Gene expression, which lies at the basis of protein production, requires "reading" DNA, for which DNA needs to be temporarily unwrapped. Detailed studies, and especially visualizations, of DNA–histone and nucleosome dynamics are crucial for a better understanding of DNA unwrapping and related processes.
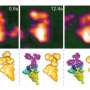
Mikihiro Shibata from Kanazawa University and colleagues have now succeeded in making video recordings of the nucleosome dynamics of H2A.Z, a histone variant associated with various biological processes. The videos reveal the spontaneous sliding of H2A.Z nucleosomes on a substrate. The work is published in the journal Nano Letters.
Histone variants, such as H2A.Z, differ from the canonical forms (like H2A) encountered in stable nucleosome packaging. They form unstable nucleosomes with particular biological functions; H2A.Z is believed to play a role in early embryonic development and stem cell differentiation. The dynamics of the H2A.Z nucleosome under physiological conditions are mostly unknown. Shibata and colleagues used high-speed atomic force microscopy (HS-AFM) to investigate H2A.Z nucleosome dynamics, as the method is a powerful nanoimaging tool for visualizing molecular structures and their dynamics at high spatiotemporal resolution.
To observe DNA–histone dynamics in HS-AFM experiments, the nucleosome needs to be put onto a substrate. The DNA should adsorb easily to the substrate, but at the same time, substrate–DNA interactions should still be weak enough to avoid suppressing dynamical processes. The scientists therefore prepared substrates by putting pillar[5]arenes onto a mica surface. The pillar[5]arenes, molecules with a pentagonal tubular structure, form a thin film on the mica and provide the ideal surface for nucleosome dynamics observations.
The researchers looked at the time evolution of a system consisting of a nucleosome particle put on a DNA strand. Experiments with canonical H2A histones confirmed the stability of H2A nucleosomes: no significant changes over time were observed. Observations for H2A.Z histone variants showed a different picture, however. HS-AFM with a time resolution of 0.3 s revealed sliding events, in which a nucleosome particle slides along the DNA strand.
The findings of Shibata and colleagues may lead to a better understanding of the biochemical mechanisms behind gene expression. "The single-molecule imaging by HS-AFM presented here could help unveil the relationship between nucleosome dynamics and gene regulation … in the near future," say the researchers.
High-speed atomic force microscopy
The general principle of atomic force microscopy (AFM) is to make a very small tip scan of the surface of a sample. During this horizontal (xy) scan, the tip, which is attached to a small cantilever, follows the sample's vertical (z) profile, inducing a force on the cantilever that can be measured. The magnitude of the force at the xy position can be related to the z value; the xyz data generated during a scan then result in a height map providing structural information about the investigated sample.
In high-speed-AFM (HS-AFM), the working principle is slightly more involved: the cantilever is made to oscillate near its resonance frequency. When the tip is moved across a surface, the variations in the amplitude (or the frequency) of the cantilever's oscillation—resulting from the tip's interaction with the sample's surface—are recorded, as these provide a measure for the local "z" value. AFM does not involve lenses, so its resolution is not restricted by the so-called diffraction limit as in X-ray diffraction, for example.
HS-AFM results in a video, where the time interval between frames depends on the speed with which a single image can be generated (by xy-scanning the sample). Researchers at Kanazawa University have in recent years developed HS-AFM further, so that it can be applied to study biochemical molecules and biomolecular processes in real-time. Mikihiro Shibata and colleagues have now applied the method to study nucleosome dynamics, revealing a sliding process of nucleosome particles along a DNA strand.
More information: Shin Morioka et al, High-Speed Atomic Force Microscopy Reveals Spontaneous Nucleosome Sliding of H2A.Z at the Subsecond Time Scale, Nano Letters (2023). DOI: 10.1021/acs.nanolett.2c04346
Journal information: Nano Letters
Provided by Kanazawa University