This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New study decodes one of the world's fastest cell movements
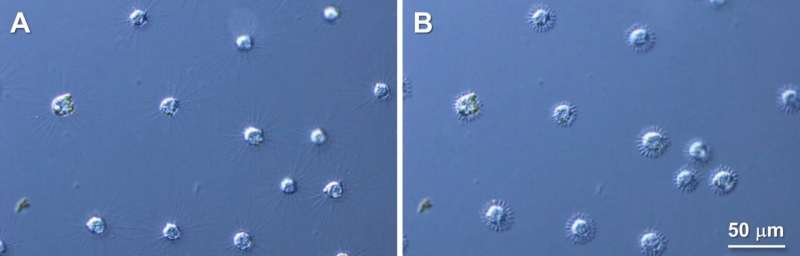
Heliozoan axopodia are important for their motility. However, the underlying mechanism of their axopodial contraction has remained ambiguous. Recently, researchers from the Okayama University reported that microtubules are simultaneously cleaved at multiple sites, allowing the radiating axopodia in a heliozoan, Raphidocystis contractilis, to disappear almost instantly.
They have now identified the gene set and proteins involved in this microtubule disruption. This research can help develop a method to detect water pollution and evaluate the efficacy of new anticancer drugs.
Raphidocystis contractilis belongs to Heliozoa, a group of eukaryotes commonly found in fresh, brackish, and sea water. The organisms of this group have finger-like arms—axopodia—which radiate out from their body, giving them a sun-like appearance. Hence, they are also known as "solar worms."
Each axopodium is composed of the proteins, alpha-beta tubulin heterodimers, which form filaments called microtubules. R. contractilis can withdraw its axopodia extremely fast in response to external stimuli. However, the mechanism underlying this rapid arm shortening remains a mystery.
To this end, a team of researchers including Professor Motonori Ando, Dr. Risa Ikeda (both from the Laboratory of Cell Physiology) and Associate Professor Mayuko Hamada (from the Ushimado Marine Institute), of Okayama University, Japan, explored the mechanism involved in one of the fastest cell movements in the living world.
So, where did it all begin? Professor Ando says, "Recently, a wide variety of heliozoans have been discovered in various hydrospheres in the Okayama Prefecture, making it clear that several species of sun worms inhabit the same environment. We are trying to unravel the mysteries around these protozoans and gradually expand the horizons of our knowledge."
The authors started their investigation by immunolabelling the tubulin protein and observing its movement before and after axopodial contraction. They found that before shortening, tubulins were arranged systematically all along the length of the axopodia, but after axopodial withdrawal, those swiftly accumulated at the cell surface.
This led them to believe that during the rapid axopodial withdrawal, the microtubules broke down into tubulin instantly. However, microtubule degradation is generally not a rapid phenomenon; it progresses rather slowly.
How then, could R. contractilis achieve this change so quickly?
The researchers hypothesized that this was possible if the microtubules split at multiple sites simultaneously. To validate their hypothesis, the authors set out to find the proteins and genes involved in the instant cleavage of microtubules in R. contractilis. Their findings were published online in the Journal of Eukaryotic Microbiology.
The researchers performed de novo transcriptome sequencing (analysis of the genes expressed at a particular time in a cell) and identified close to 32,000 genes in R. contractilis. This gene set was most similar to that found in protozoans (which are single-celled organisms), followed by metazoans (multicellular organisms with well-differentiated cells; this includes humans, and other animals).
Homology and phylogenetic analysis of the obtained gene set revealed several genes (and their corresponding proteins) involved in microtubule disruption. Among these, the most important ones were katanin p60, kinesin, and calcium signaling proteins. Katanin p60 was involved in controlling the axopodial arm length.
Several duplicates of kinesin genes were found. Among the identified kinesins, kinesin-13, a major microtubule destabilizing protein, was found to play an important role in the rapid contraction of axopodia. Calcium signaling genes regulate the entry of calcium ions into the cell from its surroundings and the induction of axopodial withdrawal.
The researchers also noticed a lack of genes linked with flagellar formation and motility, indicating that the axopodia of R. contractilis have not evolved from flagella. Although many genes remain unclassified, the newly established gene set will serve as a reference for future studies aiming to understand the axopodial motility of R. contractilis.
Heliozoan axopodia can function as a sensitive sensor. They can detect minute changes in their environment, e.g., the presence of heavy metal ions and anticancer drugs.
Discussing their vision for the future, Professor Ando says, "We believe that the axopodial response of heliozoa can be used as an index to develop temporary detection and monitoring devices for environmental and tap water pollution. It can also be used as a novel bioassay system for the primary screening of novel anticancer drugs. In the future, we plan to continue to work together as a team to enhance basic and applied research on these organisms."
More information: Risa Ikeda et al, De novo transcriptome analysis of the centrohelid Raphidocystis contractilis to identify genes involved in microtubule‐based motility, Journal of Eukaryotic Microbiology (2022). DOI: 10.1111/jeu.12955
Journal information: Journal of Eukaryotic Microbiology
Provided by Okayama University