High-resolution microscopy for analysis of protein complexes
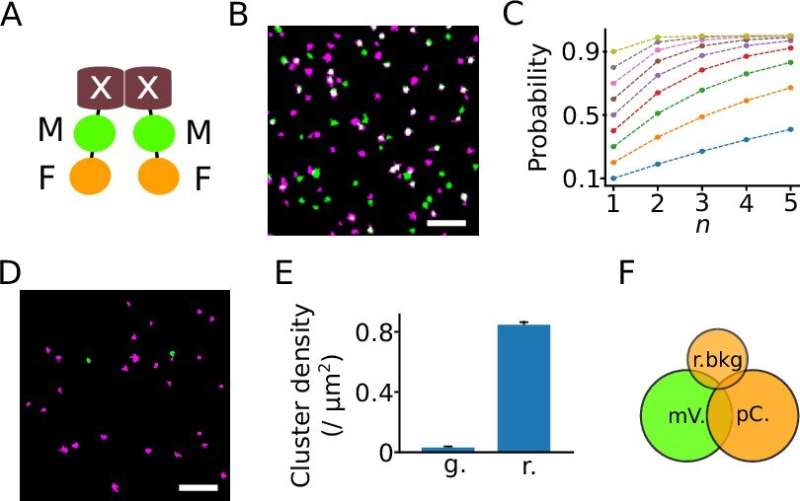
Researchers at Forschungszentrum Jülich and the Berlin Institute of Health at Berlin's Charité Hospital have developed a novel method for determining the number of subunits within protein complexes. The method is a further development of "super-resolution" single molecule localization microscopy (SMLM), whose developers were awarded the Nobel Prize in Chemistry in 2014. The new technique allows researchers to analyze the composition of protein complexes in intact cells.
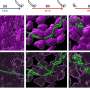
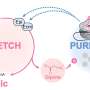
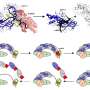
The new method is based on classical SMLM, but here proteins are labeled not with one, but with two different fluorescent proteins. This special feature has led to the name DCC-SMLM, while "DCC" stands for "dual-color colocalization". The degree of overlap ("colocalization") of the two color signals is used to calculate the average number of subunits per protein complex.
Proteins are the basic building blocks of life. They are responsible for the structure and function of cells and are involved in virtually every task of the organism.
However, many proteins do not function alone, but as subunits of larger protein complexes. Knowing how many subunits make up such a protein complex is important for understanding disease-causing dysfunctions. There are a number of genetic diseases, which are associated with a disturbed assembly of protein complexes, for example defects in the assembly of ion channels in cardiac arrhythmias, epilepsies or renal dysfunction.
The application of the classical SMLM method is limited by its sensitivity to background signals. These background signals are unavoidable within intact biological samples and must be isolated from specific signals by the biological sample. This requirement makes studying proteins in intact cells often difficult.
The new DCC-SMLM technique is significantly less sensitive to such interfering signals and allows accurate counting even when using less efficient fluorescent markers. Thus, DCC-SMLM permits studying protein complexes in the cell membrane of intact cells.
In their work, the researchers were able to elucidate the composition of complexes responsible for the transport of the neurotransmitter glutamate in nerve cells. The scientists also showed that protein complexes of the so-called SLC26 family consist of two subunits even in intact cells.
These are found, for example, in the intestine, in the kidney and in hair cells in the inner ear, where they function as motor proteins and endow hearing with extraordinary sensitivity. Previous studies had been contradictory, predicting four subunits in intact cells but only two subunits per complex in purified proteins.
The research was published in eLife.
More information: Hua Leonhard Tan et al, Determination of oligomeric states of proteins via dual-color colocalization with single molecule localization microscopy, eLife (2022). DOI: 10.7554/eLife.76631
Journal information: eLife
Provided by Forschungszentrum Juelich