Genetic analysis key to understanding Legionella risk, study finds
Routine sampling of water supplies and genomic sequencing to determine the entire genetic makeup of Legionella bacteria could play a key role in identifying the source of Legionnaires' disease outbreaks, research suggests.
The measures could also inform public health measures to limit the spread of infection, according to a genomic study of the Legionella bacteria that causes the disease. The research appears in The Lancet Microbe.
Improved testing is crucial, researchers say, because the disease—a severe form of pneumonia—is contracted by inhaling the bacteria in aerosols—tiny droplets—of contaminated water . Outbreaks are often linked to water systems in hotels, cruise ships and hospitals, and also within the community.
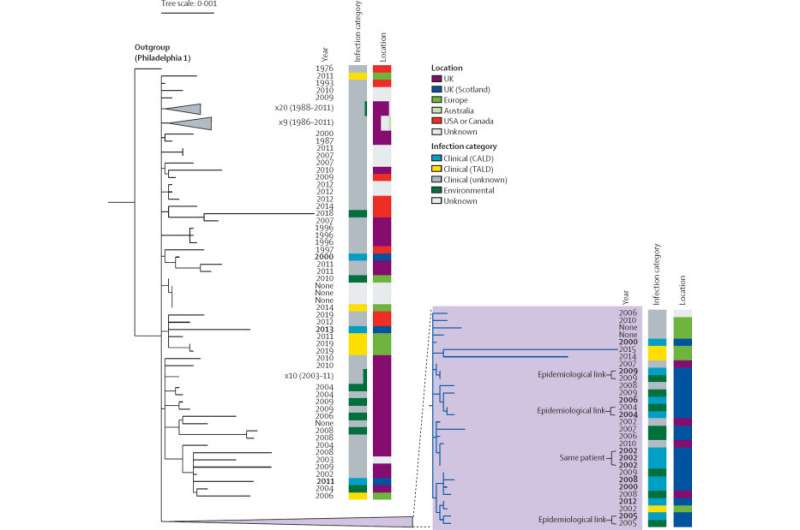
A team from the Universities of Edinburgh and Glasgow, Public Health Scotland and the Scottish Legionella Reference Laboratory conducted a comparison of the entire genome or genetic code of more than 3,000 Legionella bacteria samples found in patients and water sources from Scotland and around the world.
The results revealed a new level of detail and fresh insights into the characteristics of Legionella bacteria and its transmission in Scotland.
From analysis of the genetic code, researchers found that Legionella infections following travel were often closely related to other variants from the same U.K. or international travel destination.
The analysis found that nearly one third of infections in Scotland acquired in the community—not hospital or travel associated—were caused by a single variant. The team say this variant should be closely monitored.
Distinct variants of the bacteria were found in some hospitals over a long period of time—up to 17 years in some instances—which suggests that the bacteria persisted or were repeatedly introduced into hospital water systems.
The team also found that there had been no reported cases of Legionnaires' disease in any Scottish hospital in the past 10 years. This is likely due to effectiveness of new control measures introduced during this time, experts say.
The study was carried out by a team of experts including Ph.D. student Jamie Gorzynski, who holds a joint Edinburgh/Glasgow University Ph.D. Studentship in Precision Medicine.
Professor Andrew Smith, co-author on the study from the University of Glasgow said, "As head of the Scottish Legionella Reference Laboratory it has been a privilege to be part of this study which is a fantastic exemplar of teamworking involving National and Internationally recognized experts.
"We have been able to have a more detailed understanding of the population biology of Legionella pneumophila informing future sampling strategies and also a framework for investigating sources of future outbreaks as part of public health control measures in Scotland."
Professor Ross Fitzgerald, lead researcher from University of Edinburgh's Roslin Institute, said, "Our study indicates that regular sampling of water systems and genome sequencing of Legionella could be used to identify the source of new pathogenic variants before they become a clinical problem."
More information: Jamie Gorzynski et al, Epidemiological analysis of Legionnaires' disease in Scotland: a genomic study, The Lancet Microbe (2022). DOI: 10.1016/S2666-5247(22)00231-2
Provided by University of Glasgow