Bacterial quorum quenched by bacterial enzyme
Bacteria produce slimy biofilms to coat and protect themselves and make them less susceptible to antimicrobial drugs and chemical cleaning products. The biofilms have implications for health and various industries, as they can cause fouling, corrosion and bacterial contamination.
Now, scientists have made an enzyme that effectively breaks down the signaling molecules used by bacteria to tell each other to produce the biofilms needed to encompass their community.
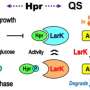
The enzyme has its roots in a 2016 discovery by KAUST researchers, when they identified seven "quorum-quenching" genes from bacteria extracted from Red Sea sediment. These genes code for enzymes that break down the quorum-sensing molecules used by bacteria to communicate with each other.
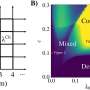
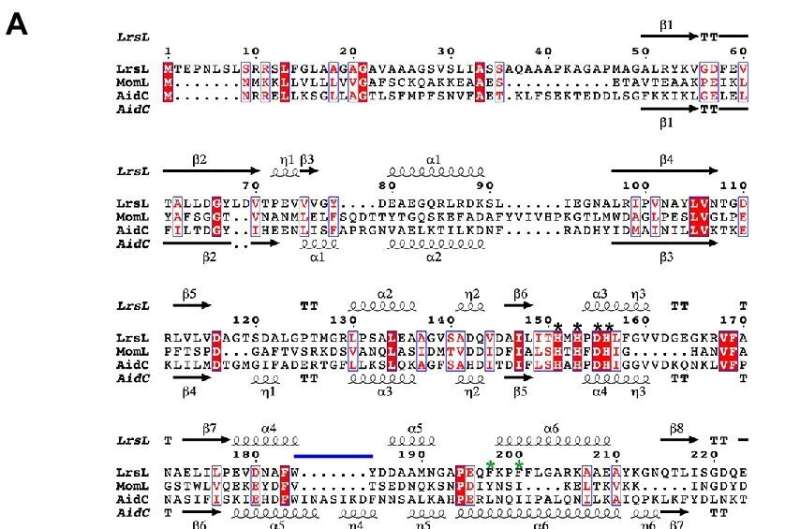
In the current study, an interdisciplinary team of scientists used 3D protein modeling to predict the structures of the enzymes coded by the seven genes. Based on their analyses, the team modified one of the genes to produce an enzyme they called LrsL, named after the Red Sea (rs) Labrenzia (L) bacterium and its lactonase (L) enzymatic activity. LrsL specifically breaks down a class of quorum-sensing molecules called acyl-homoserine lactones (AHLs).
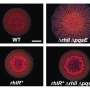
Structural and functional analyses of the enzyme demonstrated that it very effectively suppressed biofilm formation by the bacterium Pseudomonas aeruginosa, which is well known for causing hospital-acquired infections. "In fact, LrsL had the highest catalytic activity reported for any quorum quenching enzyme reported to date," says molecular biophysicist Stefan Arold. This exceptional activity is likely due to an unusual pocket in the enzyme that allows LrsL to bind extremely strongly to a broad range of AHL molecules.
Also, even though LrsL lost its structure and function when heated above 50 degrees Celsius, it was able to regain both upon cooling, even after prolonged heat treatment at 120 degrees Celsius. This characteristic makes this enzyme a promising candidate for applications in clinical and industrial settings.
"Current chemical antifouling and biofilm treatments are either toxic to the environment or involve antibiotic use, which can give rise to antimicrobial resistance," says biotechnologist Afaque Momin. "Quorum-quenching enzymes show promise for overcoming some of these challenges. LrsL has exceptional efficacy, stability and strong action on biofilms, making it a potent choice to counter biofilm formation in a variety of settings."
Further studies are needed to confirm these results before LrsL can be considered for commercial applications. In the meantime, the team will continue to test their Red Sea sediment samples for even faster acting and more stable quorum-quenching enzymes.
The research was published in Frontiers in Microbiology.
More information: Zahid Ur Rehman et al, The exceptionally efficient quorum quenching enzyme LrsL suppresses Pseudomonas aeruginosa biofilm production, Frontiers in Microbiology (2022). DOI: 10.3389/fmicb.2022.977673
Journal information: Frontiers in Microbiology