Researchers unravel cell biology through artificial intelligence
For our cells to proliferate, differentiate or migrate, the nucleus needs the help of its cytoskeleton, the scaffold surrounding the nucleus which provides cells with shape and solid structure. The disruption of this strong coupling, such as the dislocation of the nucleus from its cytoskeleton, is usually a symptom of disease in the body.
However, this relationship between the placement of the nucleus and cytoskeleton organization has never been demonstrated before due to the difficulty in being able to mathematically define the intricate design of the cytoskeleton.
Using conventional scientific methods, a scientist would need to first determine the parameters needed to define and measure the system that is being studied. This human interpretation of reality allows for the measuring of simple systems using well-known parameters such as size, speed and distance. However, for many complex systems, such as the mesh of fibers forming the cytoskeleton, defining the parameters that are important becomes an impossible task.
"Interpreting such complex systems is difficult because we must fit them into our interpretation of reality and its predefined measurables. With the thousands of intermingled spaghetti-like fibers, it would be humanly impossible to tell where one starts and the other ends, let alone figure out the parameters of the study," explained principal investigator Assoc Prof Fernandez from SUTD.
The researchers then decided to disentangle the issue from a completely new perspective, shifting their focus from the system, to the observer instead.
Assoc Prof Javier G. Fernandez and Ph.D. candidate Jyothsna Vasudevan from the Singapore University of Technology and Design (SUTD) collaborated with National University of Singapore and the Nanyang Technological University and successfully demonstrated the correlation between cytoskeleton organization and nuclear position by turning to artificial intelligence. Their study, "From qualitative data to correlation using deep generative networks: Demonstrating the relation of nuclear position with the arrangement of actin filaments," was published in PLOS ONE.
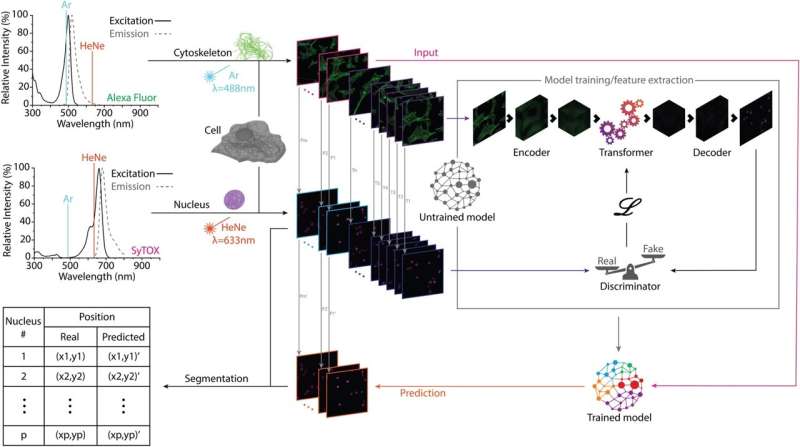
To ensure that the study's parameters would not be limited by human conceptualization, they developed a unique generative algorithm to interpret the cytoskeleton of eukaryotic cells using qualitative data, without telling the system what it was observing and how to measure it.
"We separated the information related to the nucleus and the fibers in independent databases of images, ensuring that there wasn't any information about the nucleus found in the images of the fibers, so that the system couldn't cheat. Then we trained the system to find the location of the nucleus using only information specific to fibers. To do so, the system had to take the qualitative data and figure out on its own if there was a relation between the organization of the fibers and the position of the nucleus. This forced the program to find the parameters defining the system, free from human interpretation and predefined concepts," Assoc Prof Fernandez added.
The algorithm was able to successfully predict the presence and the location of the nuclei in more than 8,000 cells, with almost half of those predictions resulting in a deviation of less than 1 μm from their exact position. This demonstrated, with astounding significance, the hypothesis of a deterministic relation between the arrangements of the actin filaments and the position of the nucleus, one of the most basic relations in cell biology. Assoc Prof Fernandez believes that this has also resulted in an epistemological outcome.
"This study has transformed the way we think about adapting our scientific research methods to allow machine learning to not just be used as a tool to analyze data, but to also interpret reality. For the inherently complex systems in biology, this will undoubtedly accelerate the next technological revolution—the 'biologization' of technology. This will enable the complexities and intricacies of biological systems to be truly unraveled and dominated using machine learning," said Assoc Prof Fernandez.
More information: Jyothsna Vasudevan et al, From qualitative data to correlation using deep generative networks: Demonstrating the relation of nuclear position with the arrangement of actin filaments, PLOS ONE (2022). DOI: 10.1371/journal.pone.0271056
Journal information: PLoS ONE
Provided by Singapore University of Technology and Design