Exploring molecular boundaries in DNA
It's important to be well organized. And this is especially true for the genome—the entirety of an organism's genetic information, also known as DNA. The genomic DNA is several meters long but has to fit inside the cell's nucleus, which has a diameter of just around 5 micrometers. In mammals, the genome houses many thousands of genes and up to ten times more enhancers, which act like switches in gene activation. Sophisticated folding allows genes and enhancers to interact in an orderly manner. This is important because for an organism to develop properly and not fall ill, thousands of genes must be activated and read correctly at the right time. This is the only way that tissues and cells can form properly.
To ensure that the dynamic folding of this complex 3D structure doesn't lead to the wrong genes being activated, the genome has molecular boundaries that bring order to the chaos. Changes in the boundaries can result in birth defects or cancer. A team led by Dr. Darío Lupiáñez, head of the Epigenetics and Sex Development Lab at the Berlin Institute for Medical Systems Biology (BIMSB) at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), has now discovered previously unknown details about these molecular barriers. The research team has published its results in Nature Genetics.
The perfect match
Much like a city, the genome is divided into large functional districts. The technical term for these genomic neighborhoods is "topologically associating domains," or TADs for short. Enhancers and the genes they activate usually occupy the same TAD—and enhancers generally can't activate genes on other TADs. "Genomic segments with characteristic properties separate the TADs from each other," says Chiara Anania, a lead author of the study. These TAD boundaries function as molecular barriers making sure that nothing goes wrong during gene expression. If these boundaries change, enhancers might activate the wrong gene or fail to find their target. It's the equivalent of a package not having the zip code on it, which causes the mail carrier to go to the wrong house or not deliver the package at all. In cells, these types of errors can lead to disease.
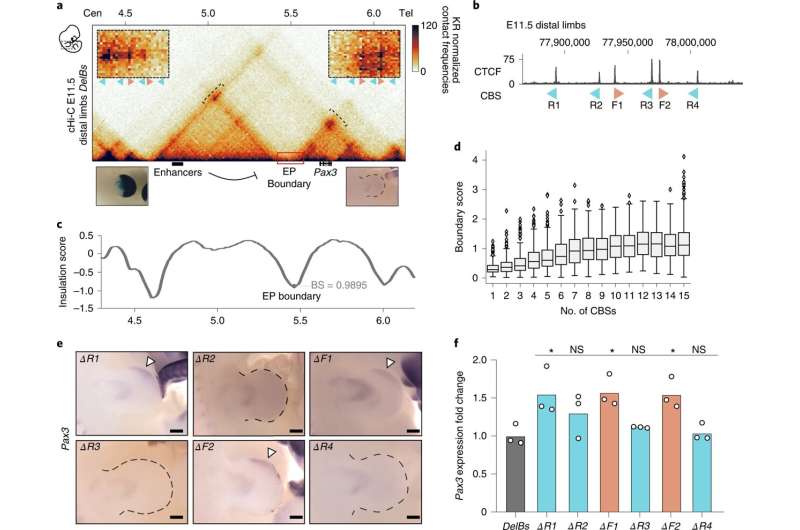
"The TAD boundaries were accurately mapped for a variety of cell types and tissues, but we didn't understand how they worked," says Anania. All they knew was that the insulating properties of the TAD boundaries were based on the binding of a protein called CTCF. This is why these relevant DNA segments have many CTCF binding sites—which essentially form the foundation for the boundary. The research team has now investigated a prototypical TAD boundary that contained six different CTCF sites. For the experiment, they used mice in which they could alter these CTCF binding sites. "We induced various mutations of these sites to understand what exactly is needed to form a functional boundary," says Anania. This allowed the team to investigate how the barriers function in an organism and how they impact gene activation—specifically in the genes responsible for limb development.
Not all boundaries are created equal
"We discovered that, although a boundary region has multiple sites capable of binding CTCF, they aren't all the same," says Rafael Acemel, also lead author of the paper. Surprisingly, some binding sites are better at insulating TADs than others. Even a TAD boundary with just two CTCF sites can offer more insulation than one with four—depending on the individual properties of each sites. The researchers also found that, to a certain degree, CTCF sites work together and can even compensate for absent sites.
"It was generally assumed that a boundary might be sufficient to prevent communication between enhancers and genes in different TADs," says Lupiáñez. "But our results suggest that the insulation shouldn't be viewed as an absolute property of TAD boundaries." When the strength of the TAD boundary changes—for instance, because fewer CTCF sites are available or they have been rearranged—this can also affect the degree of gene activation by enhancers in a neighboring TAD. The researchers found that these quantitative changes in gene expression correlated with how pronounced limb phenotypes were in their experiments.
Earlier studies showed changes at the TAD boundaries in, for instance, patients with genetic limb defects or fragile X syndrome. "In these cases, changes in the boundaries could lead to defective gene expression and cause disease," says Lupiáñez. The TAD boundaries in cancer cells are also often altered and cause unusual gene expression. Lupiáñez hopes his team's findings will help to develop new strategies to correct faulty gene expression in these types of diseases.
More information: Darío Lupiáñez, In vivo dissection of a clustered-CTCF domain boundary reveals developmental principles of regulatory insulation, Nature Genetics (2022). DOI: 10.1038/s41588-022-01117-9. www.nature.com/articles/s41588-022-01117-9
Journal information: Nature Genetics
Provided by Max Delbrück Center for Molecular Medicine