Mapping the effects of mutations in antibiotic-resistant bacteria
When a bacterium becomes more resistant to one antibiotic, it sometimes becomes more sensitive to another. To better understand this interaction, researchers from the Leiden Institute of Biology (IBL) and the Leiden Academic Center for Drug Research (LACDR) under supervision of Daniel Rozen and Coen van Hasselt mapped the effects of mutations in antibiotic-resistant bacteria. Their findings have been published in the scientific journal PNAS.
The increase and spread of antibiotic-resistant bacteria is a global problem. "It is not called a silent pandemic for nothing," says researcher Apostolos Liakopoulos. Every year, about 700,000 people die from its effects.
Resistance has a price tag
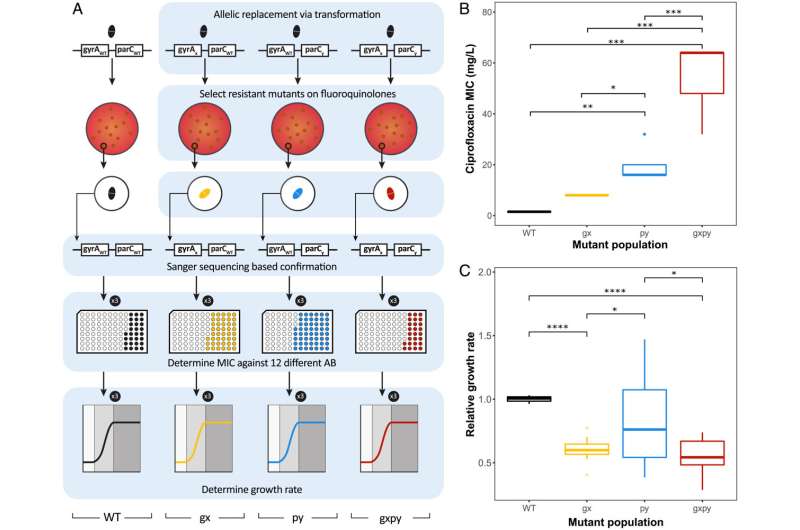
Antibiotic resistance can develop in bacteria in different ways. Liakopoulos explains: "Often it is due to mutations in genes that happen to be linked to resistance." Together with Ph.D. candidate Linda Aulin, from the quantitative pharmacology group at Leiden University, he attempted to map out the effects of these types of mutations. "Sometimes a bacterium develops resistance to one antibiotic and is therefore less able to tolerate another antibiotic," he states. This interaction is called collateral sensitivity. A phenomenon that can potentially be used to prevent antibiotic-resistance development and to combat resistant bacteria.
Interdisciplinary collaboration
Whether or not there is a question of collateral sensitivity depends on the mutation, the bacterial species and the type of antibiotics. According to Aulin, researchers often only look for interesting relationships between bacteria and antibiotics and usually stop there. "That's a shame, because you can't translate such lab results directly into a treatment suitable for real patients," she says. So the researchers joined forces. Aulin used the experimental results of Liakopoulos' research on the effects of mutations on the degree of antibiotic resistance to create computer models.
Virtual patients
Using mathematical models, Aulin infected virtual patients with bacteria. "The power of this type of simulation is that the researchers can look at all the factors that influence the course of the infection and resistance development individually. "In this way, we hope to better understand the potential effects of using collateral sensitivity to suppress resistance development within patients," says Aulin.
According to both researchers, the use of computer models is the first step in translating laboratory experiments into treatments based on collateral sensitivity. Liakopoulos: "It seems that collateral sensitivity reduces the amount of antibiotics needed to fight bacterial infections." This would be a positive development, as some antibiotics in high amounts are also harmful to humans. "This is where I think collateral sensitivity has the biggest potential," says Aulin.
Two sides of the same coin
However, more research is needed before collateral sensitivity can be used as part of a treatment. It is not very common and collateral resistance can even occur. Resistance to one antibiotic can also lead to more resistance to another. "We see this a lot with the E. coli bacterium, for example," says Liakopoulos. Therefore, it is necessary to first map the effects of mutations in multiple bacterial species. Hopefully, this will provide researchers with ammunition needed to fight the emergence of antibiotic-resistance.
More information: Apostolos Liakopoulos et al, Allele-specific collateral and fitness effects determine the dynamics of fluoroquinolone resistance evolution, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2121768119
Journal information: Proceedings of the National Academy of Sciences
Provided by Leiden University


















