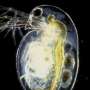
Unexpected relationships in waterflea evolution revealed by examining whole genomes
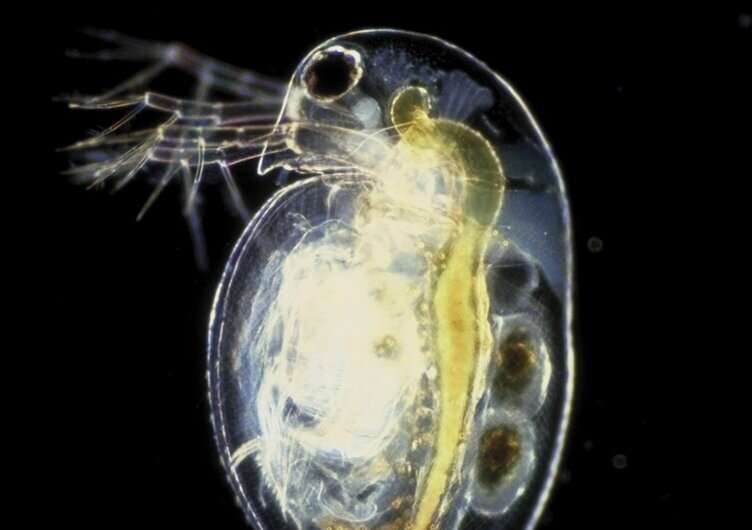
In a new study that appeared in the renowned journal Systematic Biology, scientists of Ghent University (Belgium), and the University of Basel (Switzerland) have examined the evolution of waterfleas using state-of-the-art tools. These small crustaceans of the class Branchiopoda are key zooplankton species in aquatic ecosystems worldwide. Daphnia is perhaps the best known waterflea, containing herbivorous species that are widely used model organisms in biology, yet there are also predatory lineages such as the glass waterflea (Leptodora).
The relationships between the major lineages of waterfleas, each specialized in different niches, have been debated for centuries. In addition, fossils are lacking for most groups, and the timing of major divergences remains unclear. "Despite their importance in ecology, and despite the fact that Daphnia is the aquatic equivalent of fruitflies in genetics, we understand little about the evolutionary history of the Cladocera," says Dr. Kay Van Damme, a researcher at Ghent University in Belgium.
For the first time, Van Damme and his colleagues at the University of Basel in Switzerland at the lab of Prof. Dieter Ebert, used hundreds of genes to investigate the evolution of waterfleas. More than 800 genes were used to build the first Tree of Life of all living orders of the branchiopod crustaceans, based on entire genomes. Several of these genomes were reconstructed for the first time. The scientists applied a molecular clock to the tree, calibrated by the available fossil record, to estimate when the most recent common ancestors of these lineages occurred.
"Until now, the evolutionary history of the waterfleas had been derived using a few genes or morphological traits," says Van Damme, "but relationships going back several hundred million years can become a bit blurry and new approaches were needed."
To obtain the necessary data, the team had to overcome several challenges along the way, such as extracting enough DNA from these tiny animals. For this, the researchers set up cultures, starting from one individual waterflea of each species, at the lab in Basel, and in a field station of the University of Helsinki, the Tvärminne Zoological Station (Finland).
The relationships between the lineages was not what the researchers had expected. Instead, their study revealed that major types of feeding modes (like herbivorous and predatory) and the accompanying morphologies, evolved more than once independently in these small crustaceans. For example, the predatory lineages, were assumed until now as being related, because they have similar adaptations (e.g., large eyes and spiny limbs). The authors of the study suggest that convergent evolution, resulting in similar functional traits independently (like wings in bats and birds), may be a more common mechanism in nature than generally thought. At least in waterfleas, this appears to be the case.
In addition, some lineages were estimated to go back hundreds of millions of years. The origin of the cladocerans was estimated at about 325 million years (Carboniferous), and the authors suggest that waterfleas have diversified rapidly after the end of the Permian, around 250 million years ago. The researchers state that the ancient rapid radiation may be one of the reasons why cladoceran lineages are so hard to disentangle.
According to the team, their study helps to better understand the evolution of a key group in freshwater ecosystems, but in particular they emphasize how tricky it is to examine relationships over such a large period of time. Relationships derived from morphology in ancient lineages, are not always confirmed by independent (genetic) data, as the case in their study. The techniques can be applied to reconstruct the relationships in other groups.
More information: Kay Van Damme et al, Whole-Genome Phylogenetic Reconstruction as a Powerful Tool to Reveal Homoplasy and Ancient Rapid Radiation in Waterflea Evolution, Systematic Biology (2021). DOI: 10.1093/sysbio/syab094
Journal information: Systematic Biology , PLoS Biology
Provided by Ghent University