November 17, 2021 feature
Study explores the origin of clonal dominance in excitable cell networks
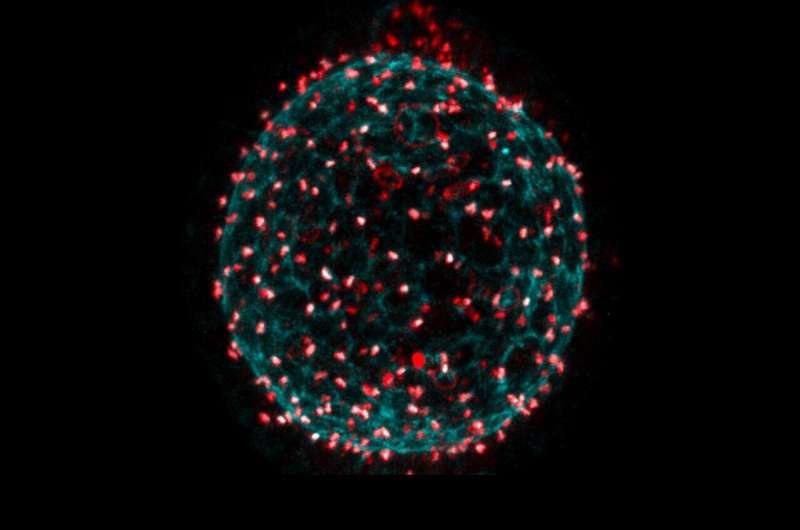
Clonal dominance is a phenomenon that occurs when descendants (i.e., clones) of one or more founder cells in an organism contribute disproportionally to the system's final structure as the tissue grows. This phenomenon is associated with numerous biological processes, including bacterial growth and the genesis of tumors. While numerous studies have investigated clonal dominance, its origin is still is poorly understood.
Researchers from Princeton University, the Flatiron Institute in New York, and Jožef Stefan Institue in Slovenia have recently carried out a study investigating clonal dominance in the egg chamber of the fruit fly Drosophila melanogaster, a biological system in which the collective growth dynamics of cells can be easily reconstructed. Their paper, published in Nature Physics, suggests that clonal dominance can emerge spontaneously in excitable cell networks.
The fruit fly egg chamber is a multicellular system made up of a cluster of 16 germ cells, encased by a layer of epithelial cells. The cells in both these types of tissues are connected by ring canals, stabilized intercellular bridges that connect the two daughter cells arising from a division. Therefore, scientists can reconstruct cell lineage trees simply by tracing the ring canal connections between cells.
Lineage tracing techniques, a set of tools used to identify and track cell populations in vivo, have advanced significantly over the past few years. These are particularly promising for studies examining complex, 3D and heterogeneous tissues, containing 10s or 100s of thousands of cells. The egg chamber epithelium, which has fewer cells that form a single layer, is much simpler and is therefore well suited for lineage tracing.
"In the egg chamber epithelium, descendants remain, for the most part, connected to each other through ring canals, making cell lineage tracing more straightforward," Jasmin Imran Alsous and Jan Rozman, two of the researchers who carried out the study, told Phys.org. "In addition, this system has a relatively clear start and end points for cell divisions: starting with ~50 cells and ending with ~1,000."
A series of past studies led by Lynn Cooley at Yale Medical School and Alan Spradling at Howard Hughes Medical Institute, identified the various ring canal proteins, as well as the number and location of founding stem cells that give rise to the epithelium, respectively. The recent work by Imran Alsous, Rozman and their colleagues built on these previous findings.
"In our previous work, we focused almost exclusively on the cluster of 16 germ cells where ring canals are quite large, growing from ~1 micron to ~10 microns in the larger egg chambers, and allowing passage of most RNAs, proteins and organelles," Imran Alsous said. "Using 3D imaging, image processing and theory, we showed that as these cells grow, they do so unequally and that the resulting differential growth pattern could be rationalized by accounting for polarized transport through the ring canals."
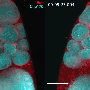
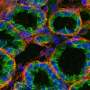
In a subsequent paper, Imran Alsous and her colleagues also investigated how these germ cells pack within an enclosure, given the topological constraints they uncovered. In the summer of 2018, they received a vial of fruit flies from another researcher, in which a protein that localizes to the ring canals was labeled, so that ring canals could be seen through fluorescence microscopy. They thus decided to investigate the extent of cell connections in the epithelium.
"In the germ cells, ring canals have been shown to be important for synchronizing the divisions that give rise to the 16-cell cluster—going from 1 cell to a 2-, 4-, 8-, and eventually a -16 cell cluster," Imran Alsous explained. "Previous studies had already shown that ring canals in the epithelium are actually smaller than those connecting the germ cells, on the order of ~300 nm, but are nonetheless large enough to allow protein equilibration between connected cells through diffusion."
Using the sample they received from their colleagues, the researchers decided to trace cell connections in the epithelium in egg chambers at various stages of development, where epithelia had anywhere between ~50 cells and ~1000 cells. Their study was conducted largely out of curiosity.
"When we first started this project, in the summer of 2018, I was set to leave Princeton relatively soon," Imran Alsous said. "My co-author, Jan, was on a summer internship at Princeton."
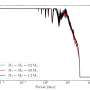
When they first started tracing cell connections in their sample, the researchers quickly realized that connected cells formed clusters that varied greatly in size. While some of these clusters contained a few or a dozen cells, others could contain hundreds of cells. The team expected to find a certain degree of variation in the size of cell clusters, however the differences they observed were far more pronounced than they anticipated. Their observations suggest that while some cells divided often, others rarely did.
After they observed large clusters of cells, Imran Alsous, Rozman and their colleagues set out to explain the origin of these clusters. Previous studies gathered valuable insight that informed their efforts, specifically delineating two facts about the fruit fly egg chamber epithelium. Firstly, studies found that in this system, the ring canals connecting cells are large enough for proteins to move through them and cross between cells. Secondly, researchers observed cell divisions in adjacent cells more often than one would expect to observe if cell division was randomly initiated.
"Combining these facts, we came upon the hypothesis that dominance could emerge because cell divisions are coupled in the epithelium (i.e., when a cell starts to divide, mitosis promoting factors could diffuse through ring canals into adjacent cells, inducing them to also divide)," Imran Alsous and Rozman said. "To test this hypothesis, we developed a mathematical model for the growing egg chamber epithelium, based on an older model that notionally represented the spread of forest fires."
The original version of the 'forest fire' model adapted by Imran Alsous, Rozman and their colleagues shows spaces on a grid that can either be filled by a normal tree, a burning tree or nothing (i.e., they previously contained a tree that burned down). The trees could catch fire either spontaneously (representing, e.g., a lightning strike) or from an adjacent burning tree. Empty spaces in the grid, on the other hand, could potentially host a newly grown tree.
In the adapted version of the model devised by the researchers, the tree grid was replaced by a network of cells connected by ring canals. Trees corresponded to a cell that can divide, burning trees to cells that were in the process of dividing, and empty spaces to daughter cells that resulted from a cell division, which are in turn not yet capable of dividing.
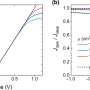
"Cells could divide either spontaneously or be induced to divide by a linked dividing cell, reminiscent of a fire jumping between trees," Imran Alsous and Rozman said. "Unlike a traditional 'forest fire' model, each new division also added a new node (i.e., the new cell emerging from the division), thereby growing the network. Using this representation of the growing epithelium, we showed that coupling of cell divisions could indeed lead to the sizes of dominant clones observed in the egg chamber, capturing their statistics and dynamics of their emergence."
The study offers new insight about clonal dominance in the fruit fly egg chamber epithelium. Most notably, it outlines a possible explanation for the emergence of dominant clones resulting from cell division coupling.
In addition, the findings offer additional evidence for the excitable nature of cell cycle and gene regulatory processes. The excitable nature of these processes, already highlighted by past studies, was also previously confirmed by both genetic and biochemical experiments.
"Our opinion is that the study almost opens as many questions as it answers," Imran Alsous and Rozman said. "For example, while it shows a possible means for the emergence of dominant clones, it is not clear if and how clonal dominance and division coupling confers an advantage to the developing system. Speculatively, it could result in faster growth or play a role in tissue scale coordination such as cell divisions stopping at ~1,000 total cells."
In their next studies, the authors also plan to explore how topological links (i.e., the ring canals) affect the tiling of surfaces and tissue-scale dynamics, as they have previously done in a study focused on the packing of the 16 germ cells.
"The 'forest fire' model in the paper is topological in nature and it doesn't account for the spatial constraints of the tissue and how growing clusters can interact or affect each other's' growth," Imran Alsous and Rozman added. "Therefore, a much more extensive model and experimental measurements would be required to understand this."
More information: Jasmin Imran Alsous et al, Clonal Dominance in Excitable Cell Networks, Nature Physics (2021). DOI: 10.1038/s41567-021-01383-0
Jasmin Imran Alsous et al, Collective Growth in a Small Cell Network, Current Biology (2017). DOI: 10.1016/j.cub.2017.07.038
Journal information: Current Biology , Nature Physics
© 2021 Science X Network