September 20, 2021 feature
Cristae-dependent quality control of the mitochondrial genome
Mitochondrial genomes (MtDNA) comprise essential subunits of the mitochondrial respiratory chain; therefore, mutations in mtDNA can reduce cellular energy support and cause mitochondrial diseases. Researchers seek to understand how cells can secure the integrity of mtDNA across generations. In a new study now published on Science Advances, Christopher Jakubke and an international research team in biology and biophysics, demonstrated how single-celled yeast Saccharomyces cerevisiae could intracellularly distinguish between functional and defective mtDNA. The outcomes of the research can support a model that shows how the proximity between mtDNA and the proteins it encodes can create a sphere of influence to understand the functional mtDNA in cells.
Mitochondria
Mitochondria contain their own genome known as mitochondrial DNA (mtDNA) that can encode core subunits of the respiratory chain including a spectrum of proteins required for mitochondrial protein translation. As a result, multiple copies of mtDNAs are distributed throughout the mitochondrial network and therefore mutations in mtDNA can be detrimental for mitochondrial function. Researchers have sought to understand how the integrity of mtDNA can be retained for generations regardless of the high rates of mitochondrial mutation. Studies on the fruit fly Drosophila melanogaster and on mice have shown how the mutant mtDNA copies can be removed in the female germline via a process known as purifying selection. For example, researchers have proposed the contribution of mitochondrial fission to the process in fruit flies, while small mitochondrial fragments containing only one or a few mitochondrial genomes can separate the mtDNA copies from one another. Despite similar progress in mitochondrial research, many questions remain to be answered relative to the cellular mechanisms facilitating selection against mutant mtDNA. Jakubke et al. therefore explored the possibility of genetically manipulating mtDNA in Saccharomyces cerevisiae; a budding yeast model to study the process of mtDNA quality control.
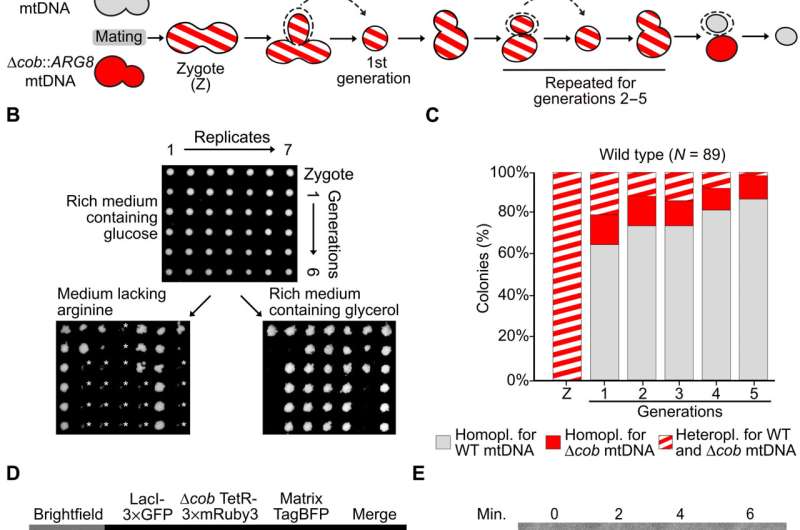
At first, Jakubke et al. tested if the single-celled S. cerevisiae could intracellularly distinguish between intact and mutant mtDNA to support the generation of young and healthy mtDNA content. During the experiment, they genetically followed the distinction of wildtype mtDNA copies from mutant mtDNA in heteroplasmic single yeast cells. To accomplish this, the team used two yeast strains of opposing mating types. The first strain harbored a wild type mtDNA and a deleted gene that typically encodes a mitochondrially localized protein known as Arg8, required to synthesize arginine. The strain grew on medium with nonfermentable carbon sources but not on medium without arginine. The next strain grew in the absence of arginine but not on nonfermentable carbon sources. Both strains maintained comparable amounts of mtDNA, which the team verified using quantitative real-time polymerase chain reaction experiments.
The experiments
Using microdissection, the team transferred single zygotes to a cell-free area on an agar plate with rich medium and glucose as a fermentable carbon source to support the growth of wild type cells or mutant mtDNA. The team continued the process of yeast cell proliferation up to five generations to respectively indicate the presence of wild type or mutant mtDNA. The method showed a strong bias for the wild type mtDNA copy. Jakubke et al. used a microscopic approach to visualize wild type vs. mutant mtDNA in the budding yeast daughter cells starting from zygotes. The results supported that the S. cerevisiae cells could distinguish between wild type and mutant mtDNA to promote the growth of healthy mtDNA content.
Selecting against mutant mtDNA in a continuous mitochondrial network
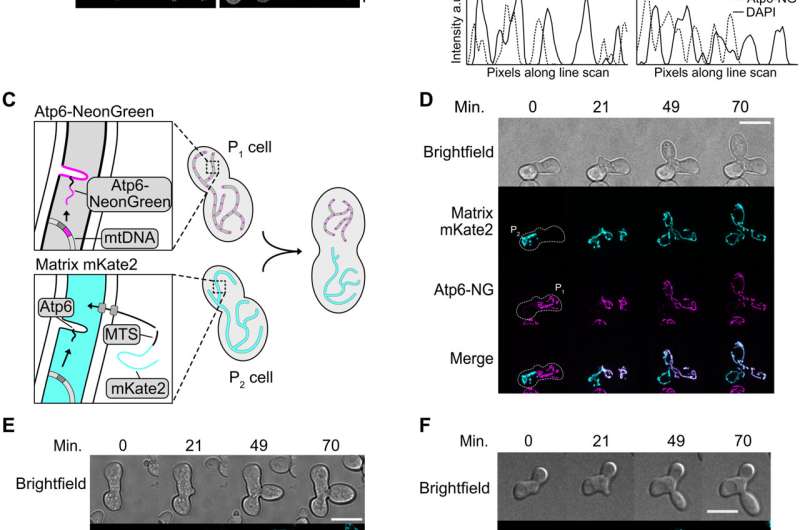
Mitochondria can form a continuous tubular network in S. cerevisiae that is constantly rearranged by fusion and fission events. For instance, dysfunctional mitochondria do not fuse with healthy mitochondria to form a continuous network and are thus kept in separate compartments. The scientists used live-cell microscopy to examine the possibility and imaged mitochondrial fusion during mating of two yeast strains. Despite pedigree mixing efforts, the yeast zygotes continued to distinguish between the mutant and wild type mtDNA to produce daughter cells with predominantly wild type mtDNA. The scientists proposed mitochondrial fission as a process to assess the selection against mutant mtDNA, but they did not identify increased mitochondrial fragmentation after mating between cells containing the wild type and mutant forms of mtDNA.
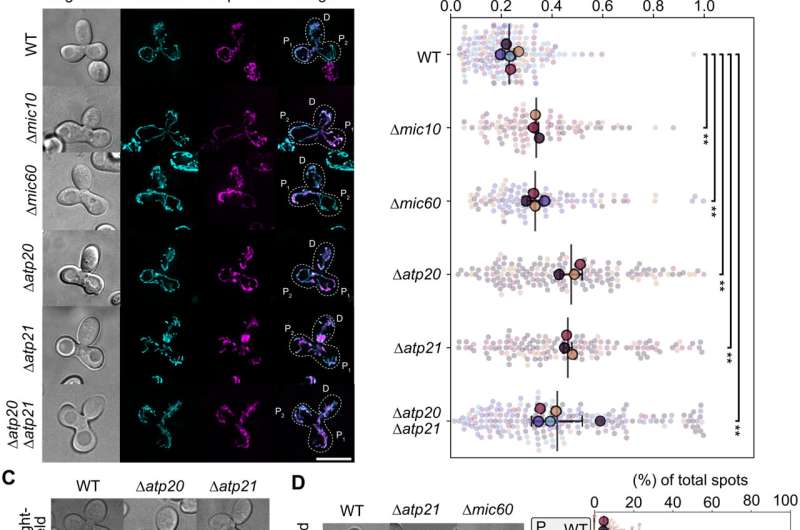
Functional compartmentalization in mitochondria
The team noted how the selection of specific wild type and mutant mtDNA in mitochondria occurred within a continuous network. They hypothesized the presence of subdomains in the mitochondrial network whose functionality is determined by copies of nearby mtDNA. To understand this, they conducted protein diffusion studies in living cells during mating experiments. The outcomes showed the equilibrium of subunits of the respiratory chain complexes throughout mitochondrial tubules when compared to proteins of the inner boundary or outer mitochondrial membranes. Further observations also highlighted the importance of cristae; the folds of the inner membrane of the mitochondria to maintain inner membrane domains and prevent mixing between gene products of different mtDNA copies. The cristae further supported the mtDNA mediated formation of mitochondrial subdomains through two mutually exclusive mechanisms. The team conducted further studies to understand the role of cristae in restricting the mobility of mtDNA. Jakubke et al. then showed how the cristae played a significant role during mtDNA quality control. For instance, the ability to choose between the wild type and mutant mtDNA severely compromised in mutants with a defective cristae architecture.
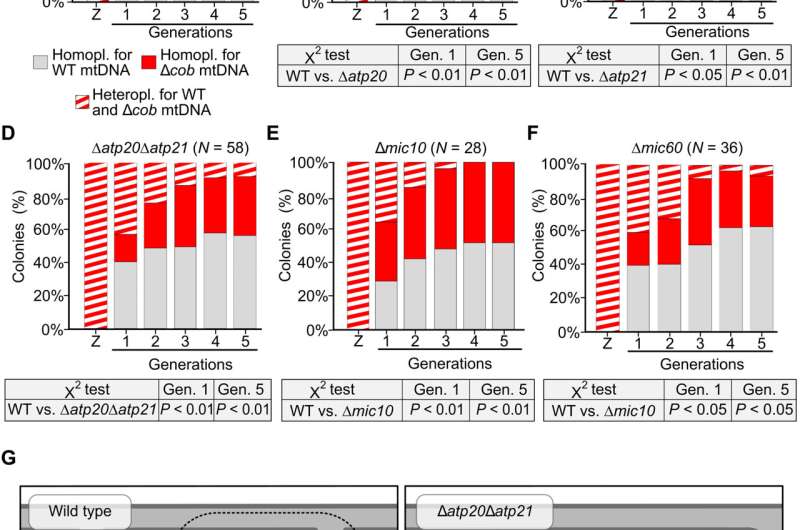
Outlook
In this way, Christopher Jakubke and colleagues showed how the unicellular S. cerevisiae promoted the generation of progeny with a healthy mtDNA population containing wild type and mutant mtDNA. The process of purifying selection occurred in a continuous mitochondrial network without mitochondrial fusion. The findings led to further investigations to understand the mechanisms cells use to detect mutant mtDNA, when compared to wild type copies, within the mitochondrial network. Based on pedigree analysis experiments, Jakubke et al. showed how mutants with defective cristae were incapable of identifying between the wild type and mutant mtDNA to support progeny with functional mtDNA. The research outcomes also supported a sphere of influence that depended on retaining the normal cristae architecture. The scientists highlighted the active presence of a purifying selection process in S. cerevisiae based on several experiments to understand the molecular mechanisms underlying the clearance of mutant mtDNA from healthy cells.
More information: Christopher Jakubke et al, Cristae-dependent quality control of the mitochondrial genome, Science Advances (2021). DOI: 10.1126/sciadv.abi8886
Jahda H Hill et al, Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant, Nature Genetics (2014). DOI: 10.1038/ng.2920
Karin B. Busch et al, Quality matters: how does mitochondrial network dynamics and quality control impact on mtDNA integrity?, Philosophical Transactions of the Royal Society B: Biological Sciences (2014). DOI: 10.1098/rstb.2013.0442
Journal information: Science Advances , Nature Genetics
© 2021 Science X Network