Clever biomolecular labeling enables identification of immune cells
Biomolecules regulate the biological functions inside every living cell. If scientists can understand the molecular mechanisms of such functions, then it is possible to detect the severe dysfunction which can lead to illness. At a molecular level, this can be achieved with fluorescent markers that are specifically incorporated into the respective biomolecules. In the past, this has been achieved by incorporating a marker in the bio-molecule by completely rebuilding it from the beginning, necessitating a large number of steps.
Unfortunately, this approach not only takes a lot of time and resources, but also produces unwanted waste products. Researchers at the Universities of Göttingen and Edinburgh have now been able to show that a non-toxic complex of the common metal manganese makes it possible to conveniently label a class of special bio-molecules known as peptides right at the last minute of their synthesis. This means the mechanism of action of these labeled peptides can be investigated efficiently. The results were published in the journal Nature Communications.
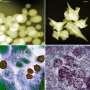
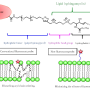
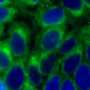
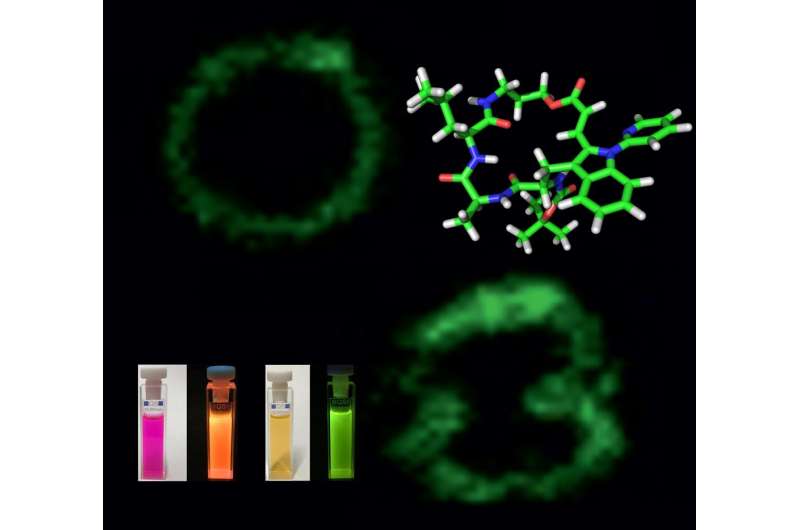
The research group developed the selective labeling of peptides and natural biological products at a late point in the series of steps necessary for synthesis by activating carbon-hydrogen bonds in tryptophan residues. This experimentally simple strategy makes it possible to efficiently access novel fluorescent pep-tides that are highly sensitive to their biological environment. This enabled the team to create a highly sensitive "rotor" with the ability to show changes in the composition of membranes of immune cells. Its fluorescence is dependent on the viscosity of the cell membrane. The researchers observed radiant fluorescence in the presence of cholesterol in the cell membrane. In this way, the rotor can be used to screen certain molecules in cells that are important for the adaptive immune system to fight infections and cancer.
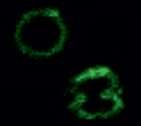
"The project demonstrates the power of combining chemical, biological and medical research, which allows the direct observation of cell-specific events," says project leader Professor Lutz Ackermann from Göttingen University. "Furthermore, the successful collaboration ensures our discoveries to have an immediate impact not only in the field of chemistry, but also in the biomedical sciences. Sharing ideas and expertise between the teams enabled a joint approach to real-life problems."

More information: Nikolaos Kaplaneris et al, Chemodivergent manganese-catalyzed C–H activation: modular synthesis of fluorogenic probes, Nature Communications (2021). DOI: 10.1038/s41467-021-23462-9
Journal information: Nature Communications
Provided by University of Göttingen