Fine-tuning metabolic gene expression to identify variants in yeast genes
Scientists have developed a more nuanced library approach to tuning gene expression in metabolic pathways. Compared to the traditional way, which leverages an all-or-nothing approach to expression, scientists can now opt for various shades of gray. The ability to fine-tune the level of gene expression allowed researchers to identify variations of essential genes in metabolic networks that were missed using traditional approaches.
This novel form of intermediate expression, when used in addition to traditional methods, can offer scientists a more colorful palette of options; beyond off and on, scientists can determine the level of gene expression by the gradations.
Metabolic engineering could lead to sustainable pathways for making high-value bioproducts including biofuels. In addition, the team says that this experiment outlines a roadmap that is transferable to other systems, expanding the technologies available for other JGI users outside the realm of bioenergy optimization.
Life is all about balance. This study, recently published in the Proceedings of the National Academy of Sciences (PNAS), offers more ways for scientists to explore the balance of gene expression and the observable characteristics in an organism.
The experiment was led by Hal Alper, professor and associate chair at the University of Texas, Austin, and included researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab).
Researchers used a library-based approach to fine-tune gene expression. They tested this new methodology in yeast (Saccharomyces cerevisiae) to assess the potential of intermediate gene expression on two energy-filled sugars used in biofuel production, glycerol and galactose. Although they are historically well-studied, Alper's team discovered new gene targets to more efficiently grow on these sugars.
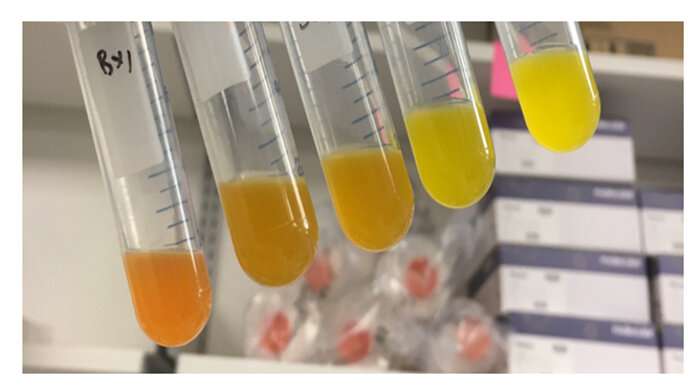
To demonstrate the versatility of this novel technique, Alper and his team also applied this method to a production-based scenario, in this case, the production of betaxanthins. They used combinations of single guide RNA, or a single RNA piece that recognizes a particular area in the DNA, to determine which targets might benefit from an intermediate level of expression. Similar to the results of the sugar-related tests, they pinpointed multiple genes that were previously missed in knock-out screens. These results show that this singular library synthesized by JGI was versatile enough for use in multiple applications.
This proof-of-concept experiment offers a new approach for fine-tuning gene expression in metabolic pathways that could be used for biofuel optimization and other applications.
More information: Emily K. Bowman et al. Bidirectional titration of yeast gene expression using a pooled CRISPR guide RNA approach, Proceedings of the National Academy of Sciences (2020). DOI: 10.1073/pnas.2007413117
Journal information: Proceedings of the National Academy of Sciences
Provided by DOE/Joint Genome Institute