April 15, 2020 report
Using DNA-like punch cards to store data
A team of researchers at the University of Illinois and the University of Texas has developed a new way to use DNA as a data storage medium—by using them like punch cards. In their paper published in the journal Nature Communications, the group describes their approach and how well it worked when tested.
Over the past several years, it has become clear that a new kind of data storage is needed—the type that is used now will not be capable of storing the massive amounts of data likely to be generated in the future. This understanding has led scientists to look for alternatives, one of which is DNA. Prior research has shown that in theory, strands of DNA should be able to hold approximately one exabyte of data per cubic millimeter. And as a bonus, it would provide a medium capable of holding data stored on it for thousands of years. But thus far, attempts to use DNA in such a capacity have fallen far short of expectations—mainly because such strands require synthesizing, a lengthy and expensive undertaking. In this new effort, the researchers have found a way to use DNA strands as a data storage medium without the need for synthesis.
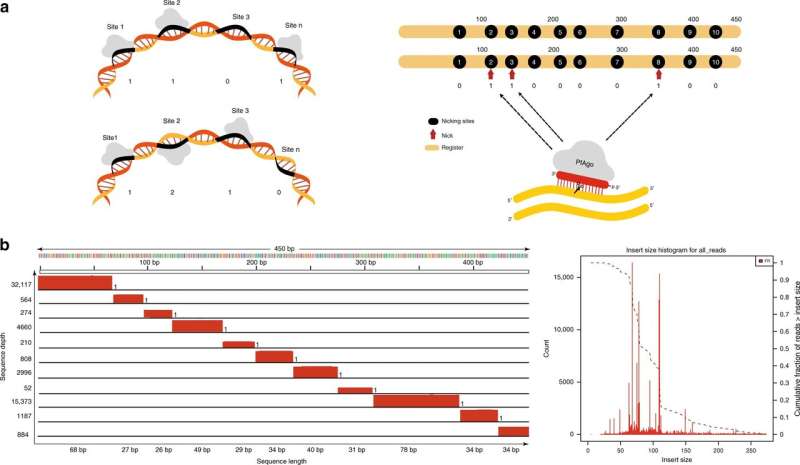
The new approach is based on old technology—punch cards: cardboard cards that represented data elements with punched holes. But instead of cardboard and holes, the new approach involves taking tiny bites out of DNA strands, leaving small nicks behind that can be used to hold information.
To accomplish this feat, the researchers heated double strands of DNA until they no longer spiraled. Doing so also led to bubble formation forcing the bases in the strands to be exposed. Then, the researchers applied single strands of DNA in 16 base lengths, which clung to the double strands. The ends of the single strands served as guides for enzymes, which cut notches by severing ties between nucleotides in the backbone rails of the original double strand. The end result was a series of notches cut into the backbone of a strand of DNA that could be used to hold data.
To test their idea, the researchers encoded parts of the Gettysburg address onto Escherichia coli DNA, and then read it back again using commercial sequencing techniques. They suggest that thus far, their approach holds great promise for resolving data storage problems of the future.
More information: S. Kasra Tabatabaei et al. DNA punch cards for storing data on native DNA sequences via enzymatic nicking, Nature Communications (2020). DOI: 10.1038/s41467-020-15588-z
Journal information: Nature Communications
© 2020 Science X Network