New study unveils the mechanism of DNA high-order structure formation
A joint research team led by Professor Ja Yil Lee (School of Life Sciences, UNIST) and Professor Ji-Joon Song (Department of Biological Sciences, KAIST) has unveiled the structure and mechanism of proteins that are highly overexpressed in cancers and associated with poor patient prognoses. The findings could speed up the discovery and development of new cancer drugs.
DNA exists in a high-order structure within cells. This structure, known as chromatin, consists of DNA wrapped around certain proteins, known as histones. The function of chromatin is to package DNA into a small volume to fit into the nucleus of a cell and protect the DNA structure and sequence.
Regulation of histone proteins allows the DNA strands become more tightly or loosely coiled during the processes of DNA replication and gene expression. However, problems may arise when histones clump together or when DNA strands intertwine. Indeed, the misregulation of chromatin structures could result in aberrant gene expression and can ultimately lead to developmental disorders or cancers.
Histone chaperones are those responsible for adding and removing specific histones during the DNA packaging process. Thus, they also play a key role in the assembly and disassembly of chromatin.
The study focused on ATAD2 (also termed ANCCA), a histone chaperone that has been implicated in nucleosome density regulation by histone H3-H4 loading or removal. It is highly overexpressed in some cancers and associated with poor patient prognoses. As a result, there has been demand for development of therapeutic agents targeting the ATAD2 protein, and some clinical trials are already underway. Yet, to date, no specific information about the structure and function of ATAD2 gene has been published.
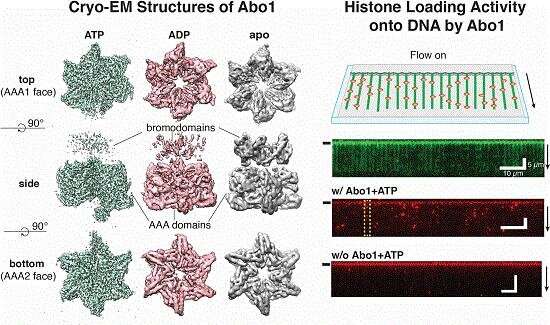
Through the use of cryo-electron microscopy (Cryo-EM), which allows direct observation of proteins in native and near-native states in atomic detail, the research team identified the structural details of ATAD2 protein. They presented cryo-EM structures of an ATAD2 family ATPase to atomic resolution in three different nucleotide states, revealing unique structural features required for histone loading on DNA, and directly visualizing the transitions of Abo1 from an asymmetric spiral (ATP-state) to a symmetric ring (ADP- and apo-states) using high-speed atomic force microscopy (HS-AFM).
Additionally, the researchers found that the acidic pore of ATP-Abo1 binds a peptide substrate that is suggestive of a histone tail. Based on these results, the scientists propose a model whereby Abo1 facilitates H3-H4 loading by using ATP.
"This study is meaningful, as it reveals the structure and mechanism of histone chaperones protein through the use of cutting-edge techniques in biophysical, such as cryo-EM," says Professor Lee. "This will accelerate the development of drug candidates, targeting ATAD2."
More information: Carol Cho, et al., "Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone," Nature Communications, (2019).
Journal information: Nature Communications