DNA-tagging strategy for early detection of disease signatures
Researchers can uncover how proteins interact with other proteins with far greater sensitivity and in a more natural context than conventional methods, thanks to an analytic technique developed by a RIKEN team. This will make it easier to discover the roles that protein-coding genes play and to diagnose conditions such as autoimmune diseases.
Many human diseases and other biological processes can only be understood by charting out networks of interactions between individual proteins. Such studies are typically performed as fishing expeditions, in which a 'bait' protein—a disease-associated molecule, for example—is exposed to mixtures of other proteins. The proteins that bind this bait can then be isolated and analyzed to reveal the partners of the disease-associated protein.
However, such methods may fail to detect relatively rare but biologically important interactions. Or they could require chemical modifications that interfere with the protein–protein interactions that normally occur within cells.
As an alternative, Junshi Yazaki of the RIKEN Center for Integrative Medical Sciences and colleagues have leveraged the power of the next-generation sequencing platforms now routinely used for genomics research.
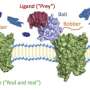
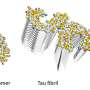
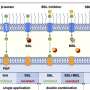
They utilized a technology called HaloTag, in which genes that encode proteins of interest are modified to include a short molecular tag. A simple chemical reaction subsequently couples the resulting protein to a unique DNA sequence, which serves as a 'fingerprint' for that protein. One can then screen a bait protein of interest against a mixed pool of tagged proteins, each with its own unique fingerprint sequence, and use next-generation sequencing analysis to assess levels of binding for these various proteins based on how highly represented their barcodes are.
This approach has several important advantages over other screening methods. "Our conjugation method is quicker and cheaper, and it can be conducted in water with just a single purification step," Yazaki says. He also notes that the HaloTag modification is relatively small, reducing the likelihood of impeding protein folding or function. Furthermore, each protein receives only a single barcode modification, allowing researchers to confidently quantify levels of interaction.
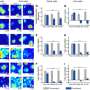
Yazaki hopes to employ this assay to detect signatures of unhealthy antibody responses in various autoimmune diseases. In an initial demonstration of their technique, the team showed that they could detect a mouse autoimmune response against a known disease-related protein with remarkable sensitivity—achieving a dynamic range that is more than 10,000 times greater than conventional diagnostic assays.
Such sensitivity could give doctors an edge in identifying such conditions and providing treatment before they progress too far. "Detection of auto-antibodies during the initial stages of a disease is critical for early diagnosis to avoid invasive testing," notes Yazaki.
More information: Junshi Yazaki et al. HaloTag-based conjugation of proteins to barcoding-oligonucleotides, Nucleic Acids Research (2019). DOI: 10.1093/nar/gkz1086
Journal information: Nucleic Acids Research
Provided by RIKEN