Scientists explain why some molecules spontaneously arrange themselves into five slices of nanoscale pie
Materials formed on vanishingly small scales are being used in medicine, electronics, manufacturing and a host of other applications. But scientists have only scratched the surface of understanding how to control building blocks on the nanoscale, where simple machines the size of a virus operate.
Now, a team of researchers led by Dongsheng Li, a materials scientist at PNNL, and collaborators at the University of Michigan and the Chinese Academy of Sciences, have unlocked the secret to one of the most useful nanostructures: the five-fold twin. Their study describing why and how this shape forms is detailed in the journal Science and was presented at the Materials Research Society annual meeting on December 5, 2019.
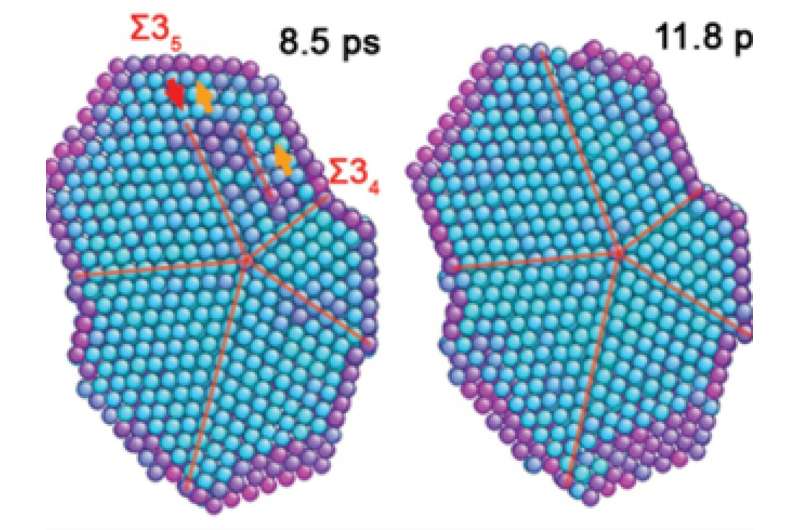
A cross section of a five-fold twin structure looks for all the world like a pie sliced into five perfectly symmetrical pieces. Nanomaterials with this structure have already been shown to have useful properties and are deployed in medical research for precisely tagging cancerous tumors for imaging and tracking, and in electronics, where they are valued for their mechanical strength.
"Natural and synthetic nanoparticles composed of five-fold twinned crystal domains have unique properties," said Li, who led the research team. "But the formation mechanism of these five-fold twinned nanoparticles has been poorly understood. For the first time, we directly observed five-fold twin formation in real time and determined the mechanism by which they form."
Ever since scientists learned how to manipulate molecules on the nanoscale, they've noted that materials tend to aggregate into certain geometric shapes: Wires, tubes, spheres, and cubes all form with little intervention. But why?
The research team used a combination of high-resolution transmission electron microscopy combined with molecular dynamics simulation techniques to probe why the structures form as they do. In most cases, nanostructures form too quickly to capture them using experimental imaging techniques. Here, the team deployed a clever strategy: they forced the molecules to move more slowly by encasing them in a molasses-like organic matrix, and they observed more than 200 formation events to capture all of the key steps of the process. They discovered two different mechanisms for forming five-fold twinned nanostructures, both of which are shaped by the accumulation and elimination of strain toward an ideal shape that eliminates all strain.
"The mechanism we worked out is a common pathway for crystal growth that occurs widely in diverse systems like metals, semiconductors, organics, and biomineral phases," said Li. "Therefore, what we learned from our observation may generalize to a wide range of materials."
Now that the researchers have captured the basic forces shaping the structure, they hope to be able to guide more kinds of materials into this very useful pie shape.
"We hope to enable the design of nanostructures with controlled size and morphology, and to tailor their properties," Li said.
More information: Miao Song et al. Oriented attachment induces fivefold twins by forming and decomposing high-energy grain boundaries, Science (2019). DOI: 10.1126/science.aax6511
Journal information: Science
Provided by Pacific Northwest National Laboratory