July 2, 2019 feature
Isolating intact bacteria from blood using a microfluidic monolith device
Emerging single-cell diagnostics rely on the potential to rapidly and efficiently isolate bacteria from complex biological matrices. In a recent study now published in Microsystems and Nanoengineering, Jung Y. Han and colleagues at the interdisciplinary Departments of Mechanical Engineering, Chemical Biomolecular Engineering and Bioengineering in the U.S. developed a device to isolate intact and viable bacteria from whole blood using a microfluidic, porous silica monolith. They achieved mechanical hemolysis while providing passage of intact and viable bacteria through the monoliths for size-based bacterial isolation and selective lysis. Han et al. described a process to synthesize large quantities of discrete capillary-bound monolith elements and millimeter-scale monolith bricks to integrate into microfluidic chips.
They explored the impact of monolithic morphology, geometry and flow conditions on cell lysis and flow regimes that allowed selective cell lysis and selective passage of multiple gram negative and gram positive bacteria. The technique employed by Han et al. allowed rapid sample preparation and bacterial analysis when combined with single-cell Raman spectrometry. The work provides unique sample preparation steps to support rapid and culture-free bacterial analysis for applications in point-of-care biomedical devices.
Bacteria in blood can lead to sepsis, infection of tissues and other serious medical conditions, requiring early identification of blood-borne bacteria for effective treatment. The ability to identify bacteria rapidly using point-of-care diagnostics can greatly enhance clinical potential for optimal treatment during early-stage infection. The existing gold standard for bacterial characterization is based on phenotypic cell culture analysis and requires at least 24 hours to collect samples for culture and analysis in a diagnostic and clinical microbiology lab. The existing technique is robust and inexpensive but cannot generate timely results to guide the initial stages of treatment.
In the present work, Han et al. explored microfluidic devices integrated with porous silica monoliths as simple flow-through elements for selective blood cell analysis and the intact isolation of bacteria. Monoliths are highly porous materials composed of open cell morphology with twisting paths of fluid flow. Scientists can control monolithic pore morphology via high mechanical surface stress during cell perfusion for mechanical hemolysis of blood cells, while allowing intact and viable bacteria to travel the winding flow paths for their culture-free isolation. Han et al. used the approach of selective passage for bacteria in whole blood under flow conditions for gram positive and gram-negative species, despite differences of the bacterial strains. The technique of high-throughput selective monolith lysis combined with powerful analytical methods such as Raman spectroscopy can allow culture-free analysis of bacteria in whole blood at the level of the single cell.
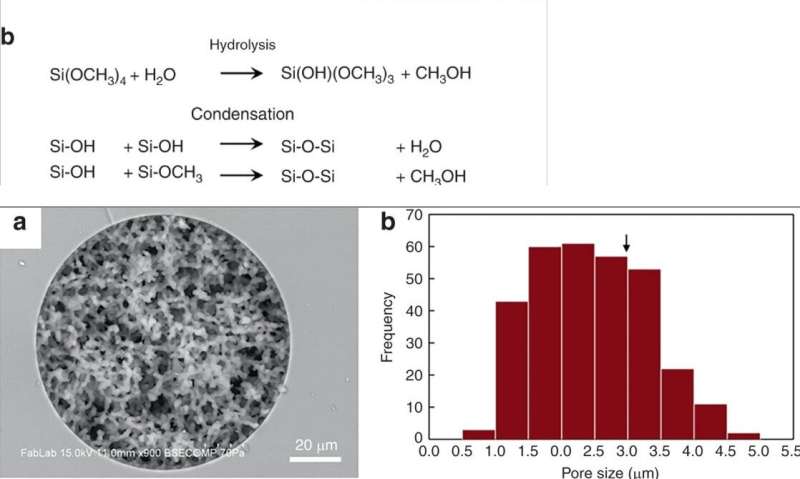
Han et al. modified previously reported silica monolith synthesis processes, followed by hydrolysis and condensation of silica to form silica glass at low temperatures. To prepare the silica monolith, the scientists used a precursor solution composed of alkyl silicates, polyethylene glycol (PEG) as a porogen, urea as a source of hydroxyl ions to minimize heterogeneity and acetic acid. When they optimized the synthetic process, the resulting monoliths were homogenous and well-anchored to the silica capillary walls. The scientists measured the thickness of the final skeletal monolith structure and calculated its permeability using high-performance liquid chromatography to control experimental conditions. To minimize intrinsic variation, Han et al. cut the resulting capillary tubes into 5 cm long segments to test permeability before use.
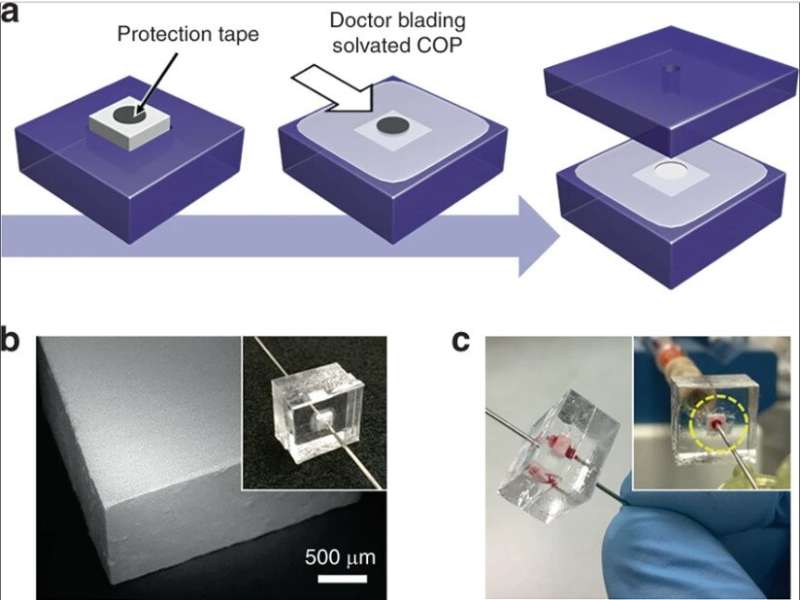
They then developed two complementary methods for low and high-throughput operation to integrate silica monoliths into microfluidic systems. To allow low throughput operation, the scientists embedded monolith-containing capillary segments within thermoplastic microfluidic chips to protect the monolith during integration. For high throughput-selective lysis they used monoliths with larger cross-sectional areas within the microfluidic devices. The complete method of fabrication yielded excellent reliability for leak-free operation during whole blood perfusion.
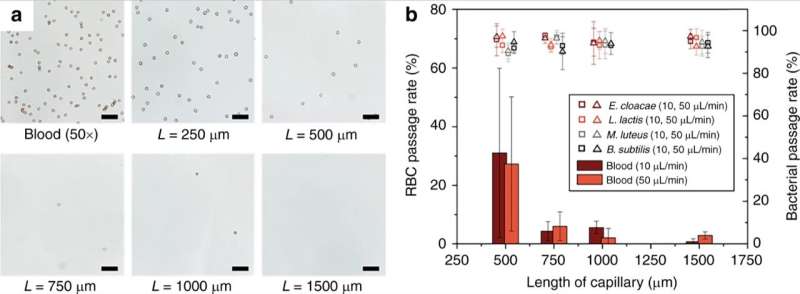
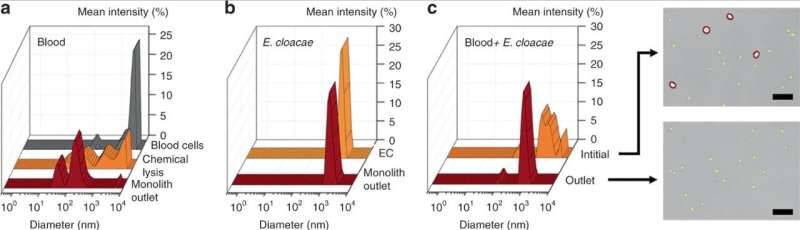
As proof-of-principle, Han et al. selected Enterobacter cloacae (gram-negative, rod shaped bacteria) to explore their efficacy of passage, alongside three gram-positive bacteria; Lactococcus lactis, Micrococcus luteus and Bacillus subtilis. During the experiments, they perfused bacterial solutions through the microfluidic monoliths with varying geometry and flow conditions to test the passage of bacteria and blood cell lysis using dynamic light scattering (DLS). For instance, the perfusion of purified E. cloacae through the monolith did not yield discernable changes in the DLS peaks, indicating the intact passage of bacteria.
The scientists showed the effect of the length of the porous monolith device on the efficiency of red blood cell lysis (RBC). The results indicated that RBC lysis efficiency increased significantly for monolith lengths above 1 mm. Han et al. also studied the fate of white blood cells (WBCs) during operation of the monolith device, the cells could not pass through the monolith without being lysed similar to RBCs. Technically, RBCs deformed to a discoid shape to pass through the monolith, which caused significantly increased membrane tension to result in RBC lysis. Comparatively, bacterial cells had similar dimensions to the monolith pores and therefore required less cell wall expansion for successful passage without rupture. The scientists optimized the parameters of the device for diverse bacteria to tolerate high levels of membrane stress without rupture. Further developments ensured the intact passage of bacteria without degradation and with viability.
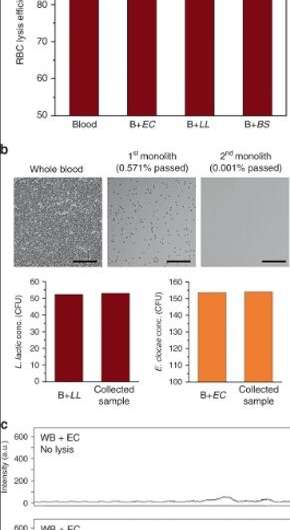
For high-throughput bacterial passage, the scientists diluted the blood in the capillary devices. However, as an alternative, they could also extend the capacity of monoliths for whole blood lysis. The devices processed more than 400 µL of whole blood spiked with bacteria before exhibiting a significant increase in back pressure, due to clogging as a result of cell lysis and also due to intact leukocytes (WBCs) trapped within the porous matrix.
To locate target bacteria, Han et al. obtained a sample deposited on to a glass slide, after it passed through the monolith-process. They conducted single-cell Raman analysis by manually scanning the optical probe across the sample. They expect the use of selective lysis technology, coupled to confocal Raman microscopy in the future to improve the process of detecting bacterial strains of interest at low concentrations in a defined location of interest.
In this way, Jung Y. Han and colleagues developed a microfluidic monolith to efficiently isolate intact bacteria with wide-ranging theranostic, point-of-care potential for clinical applications. They envision the union of confocal Raman microscopy tools that are currently largely confined to the research lab with emerging miniaturized and handheld systems to pave the way towards rapid and portable point-of-care devices.
More information: Jung Y. Han et al. Isolation of intact bacteria from blood by selective cell lysis in a microfluidic porous silica monolith, Microsystems & Nanoengineering (2019). DOI: 10.1038/s41378-019-0063-4
Susanne Pahlow et al. Isolation and identification of bacteria by means of Raman spectroscopy, Advanced Drug Delivery Reviews (2015). DOI: 10.1016/j.addr.2015.04.006
Joo H Kang et al. An extracorporeal blood-cleansing device for sepsis therapy, Nature Medicine (2014). DOI: 10.1038/nm.3640
Anna K. Boardman et al. Rapid Microbial Sample Preparation from Blood Using a Novel Concentration Device, PLOS ONE (2015). DOI: 10.1371/journal.pone.0116837
Journal information: Nature Medicine , PLoS ONE
© 2019 Science X Network