May 21, 2019 report
A better way to use atomic force microscopy to image molecules in 3-D
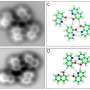
A team of researchers at Justus Liebig University Giessen has found a way to dramatically improve the images of topologically complex 3-D molecules created using atomic force microscopy (AFM). In their paper published in the journal Physical Review Letters, the group describes the simple adjustment they made to the procedure that greatly improved the resolution of AFM.
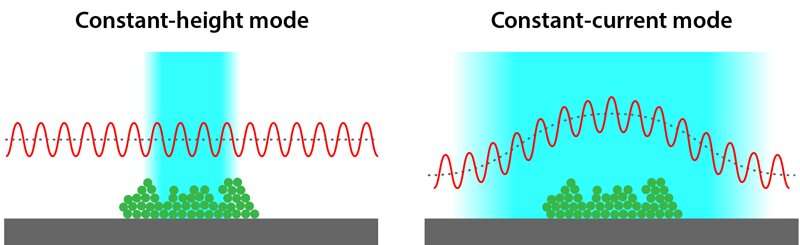
It has been nearly a decade since AFM was introduced, allowing researchers to create images of single molecules and better understand how molecules are assembled. But the technique suffers from a major deficiency—it only works on nearly flat molecules. Those molecules with more complex 3-D characteristics are visualized only partially clearly. The reason is that the tip of the sensor oscillates at a fixed distance from the molecule under study. This means only the parts of the molecule closest to the sensor are clearly visualized. Logic has suggested that the way to fix this problem is to move the tip of the probe up and down along a path that mimics the topology of the molecule. But such an approach has proven to be elusive. Tracking the hills and valleys in real time and moving the tip just the right amount has, until now, been untenable.
To overcome the problems inherent in tracking the contours of a molecule, the researchers turned to the scanning tunneling microscope (STM). It is also used to create images at the molecular level, but uses a different approach to do so. AFM uses forces from the surface under study to keep the sensor tip the proper distance for imaging—STM, on the other hand, uses the tunneling current that flows through the vacuum that exists between the sensor tip and the molecule under study. The researchers hit on the idea of using the tunneling current from STM to guide the tip of the AFM sensor tip—moving it up and down in lockstep with the contours of the molecule under study.
The researchers report that their simple adjustment resulted in images of 3-D molecules that are as sharp for complex molecules as for those that are mostly flat.
More information: Daniel Martin-Jimenez et al. Bond-Level Imaging of the 3D Conformation of Adsorbed Organic Molecules Using Atomic Force Microscopy with Simultaneous Tunneling Feedback, Physical Review Letters (2019). DOI: 10.1103/PhysRevLett.122.196101
Journal information: Physical Review Letters
© 2019 Science X Network