Cell editors correct genetic errors

Almost all land plants employ an army of cellular editors who correct errors in their genetic information. Researchers at the University of Bonn have now transferred parts of this machinery into a bacterium. Their results confirm a controversial thesis on the functioning of this widespread mechanism. They have now been published in the journal Communications Biology of the Nature Publishing Group.
One might think that the genetic machinery of higher plants was invented by a bureaucrat who likes to pick the most complicated option: Much of the plants' genetic material contains small errors. The DNA in the power plants of the plant cells, the mitochondria, is particularly affected. The plant must correct these errors, otherwise its energy supply would collapse. And it does make these corrections, but in a very complicated way: It does not improve the DNA, i.e. the actual building instructions of the mitochondria. Instead, it corrects the copies made of these instructions. This is like printing an erroneous newsletter a thousand times and then correcting the misspelled word in each of these printouts.
More than that: The editors which make these corrections are absolute specialists. They usually only recognize one specific error. Some plants therefore have 500 or more different proofreaders. "The DNA transcripts consist of RNA; we therefore call this mechanism RNA editing," explains Prof. Dr. Volker Knoop from the Institute for Cellular and Molecular Botany at the University of Bonn. "We are only just beginning to understand why it exists and how it functions in detail."
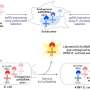
Knoop and his coworkers have at least come one step closer to answering the second question. For this purpose, they transported some editors from the moss Physcomitrella patens to the intestinal bacterium E. coli. "We wanted to find out whether they edit the bacterial RNA there," said Knoop's colleague Dr. Mareike Schallenberg-Rüdinger. "Until now it was disputed whether they can do this alone or whether they need help."
Most researchers assume that RNA editing is usually a two-step process: The editors (the so-called PPR proteins) recognize the error. To correct it, they then call on a kind of RNA 'correction fluid' - an enzyme called cytidine desaminase—for help.

RNA 'correction fluid' also works in E. coli
However, some PPR proteins have a certain sequence of amino acids at their end which are known to theoretically act as cytidine desaminase, which means they may always carry their bottle of correction fluid with them. "We were in fact able to show that this group of PPR proteins is able to edit the RNA of E. coli," said Mareike Schallenberg-Rüdinger. "So it does not need a separate desaminase to do this." However, if the scientists changed even one of the important 'correction fluid' amino acids, the PPR protein lost its ability to correct.
The researchers also succeeded in programming PPR proteins in such a way that they were able to detect specific errors. "Experiments such as these help us to better understand RNA editing," explains Volker Knoop. "The model organism E. coli also helps us in this process, as it would be much more difficult to carry out these experiments in plants."
In the medium term, the scientists also hope to find an answer to the question as to why this elaborate editorial machinery developed in the course of evolution. There are some theories on this: For example, RNA editing might enable plants to "collect" mutations. Over time, combinations of many different changes may form that would individually be harmful or even fatal, but in their sum provide the plant with a survival advantage.
The cumbersome process would therefore have an important purpose: as a playground for evolution.
More information: Bastian Oldenkott et al. Plant type Pentatricopeptide Repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli; Communications Biology, DOI: 10.1038/s42003-019-0328-3
Provided by University of Bonn