Bacteria: Protein researchers decipher resistance mechanism
Worldwide, resistance to antibiotics is on the rise. In order to understand why bacteria are becoming immune to previously well-functioning drugs, scientists are penetrating ever deeper into the molecular structure of cells. A research group at Martin Luther University Halle-Wittenberg (MLU) has now succeeded in isolating a membrane protein from the E. coli bacterium and shed light on its molecular structure. Armed with this information, they have been able to show how the bacterium manages to rid itself of the antibiotic by forcing out the drug. The paper has been published in Nature Communications.
Antibiotic resistance of bacteria is one of the most important medical issues of our time. Left unchecked, previously treatable bacterial diseases are at risk of taking such severe turns that patients might die. "This is a real threat," says Professor Milton T. Stubbs, director of the Centre for Innovation Competence (ZIK) HALOmem, where the work was conducted. According to Stubbs, who has been researching the biosynthesis of antibiotics for many years, the danger this poses means it is crucial to understand the mechanisms of antibiotic resistance.
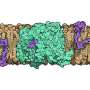
The current study is the result of work by a junior research group at ZIK HALOmem, led at the time by Dr. Mikio Tanabe. Tanabe is now an Associate Professor at the KEK Research Facility in Tsukuba, Japan. The group succeeded in isolating a membrane protein called MdfA from E. coli bacteria and were able to determine its molecular structure. The protein first had to be produced in the laboratory, isolated in its pure form and crystallised. "Dealing with sensitive membrane proteins is a very complicated process. Optimal conditions must be maintained in the laboratory so that the protein remains stable and keeps its native structure," explains Stubbs.
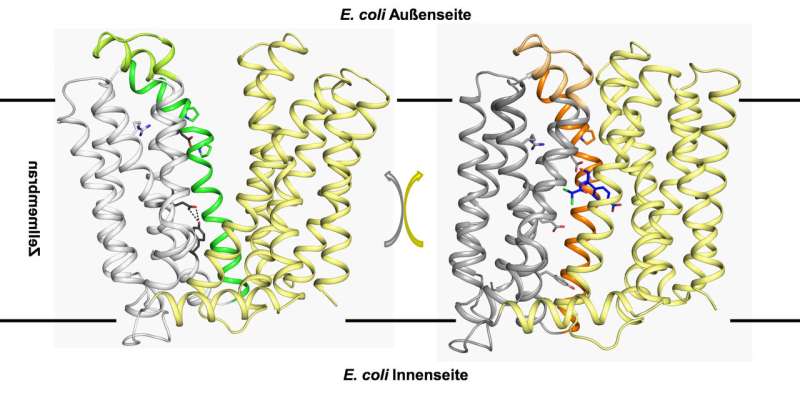
X-ray crystallography made it possible to visualise the structure of the material produced. Using this precise physical process, researchers are able to penetrate the Ångström range—one Ångström corresponds to one tenth of a nanometre, i.e. one ten billionth (10-10) of a metre, allowing researchers to work at a level at which individual atoms become visible. The ability to locate individual atoms in a molecule at this resolution holds the key to understanding how the protein works.
The process has revealed the three-dimensional structure of the membrane protein MdfA in the E. coli bacterium. The researchers from Halle utilised the results of a study that a rival group in China had recently published on the same protein and thereby succeeded in determining the mechanism that the membrane protein MdfA uses to help the bacterium become resistant.
The principle is reminiscent of a kind of pump mechanism. Although the drug is initially absorbed by the bacteria, it is ejected from the cell by MdfA before it becomes lethal to the bacteria.
"We assume that the mechanism discovered in this research applies to many other antibiotics," explains Milton Stubbs. This information will also provide the basis for later practical applications. "It is not until we understand the mechanisms of how resistances develop that we can look for solutions to prevent them."
More information: Kumar Nagarathinam et al, Outward open conformation of a Major Facilitator Superfamily multidrug/H+ antiporter provides insights into switching mechanism, Nature Communications (2018). DOI: 10.1038/s41467-018-06306-x
Journal information: Nature Communications
Provided by Martin-Luther-Universität Halle-Wittenberg