Tiny robot could be game-changer in fight against tuberculosis
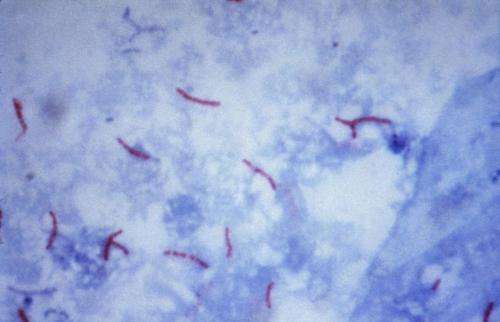
A Brock University research team has created a microscopic robot that has the potential to identify drug resistance to tuberculosis faster than conventional tests.
The World Health Organization (WHO) calls tuberculosis drug resistance "a formidable obstacle" to treatment and prevention of a disease that killed 240,000 people in 2016.
The Brock team's latest technology builds on an earlier version of the microscopic robot—called the three-dimensional DNA nanomachine—they created in 2016 to detect diseases in a blood sample within 30 minutes.
In this latest version, the team, headed by Assistant Professor of Chemistry Feng Li, re-designed the nanomachine so that it could uncover mutations in the genes found in the bacteria that causes tuberculosis.
Li says the nanomachine holds the potential to determine, within one hour, whether or not tuberculosis bacteria contain the genetic mutations that make them resistant to the basic, first-line drugs prescribed to fight tuberculosis.
The WHO says resistance occurs mostly because patients don't adhere to the strict schedule of antibiotics they need to take to get cured. The bacterial cells' genes change so that the bacteria can survive future exposures to the same antibiotics, which means a second-line treatment is then required.
It takes a while before health-care professionals and patients realize the first-line drugs aren't working, which is why quick detection of drug resistance is so crucial, Li says.
"Once you confirm there is tuberculosis infection, you have to use the diagnosis to guide the therapeutic strategy," he says. "Normal infection and drug-resistant strains require two completely different types of strategies."
Li says current testing for resistance is an arduous, time-consuming process that can take anywhere from two to six weeks and requires high-level equipment and training. In the meantime, the disease worsens in patients, who can also pass the disease along to others.
The Brock team's nanomachine consists of a 20-nanometre particle made out of gold. Short and long DNA strands are attached to the gold particle and these DNA molecules are used as building blocks to construct and operate the nanomachine.
Graduate student Alex Guan Wang used a computer simulation model to design the long strands, which are able to seek out differences in nucleotides contained within the tuberculosis bacteria's genes. A nucleotide is the basic structural unit and building block for DNA, and it's within these that mutations caused by drug resistance would be found.
The short DNA strands attached to the nanomachine carry fluorescent signal reporters.
The nanomachine is dropped into serum extracted from human blood. If the long strands detect the mutations found in specific nucleotides, the machine turns on and glows; if the sample is disease-free, the robot remains off.
Graduate student Yongya Li conducted the lab experiments. She first started the research when she was an undergraduate student.
The team's findings are contained in their paper "Simulation-guided engineering of an enzyme-powered three-dimensional DNA nanomachine for discriminating single nucleotide variants," published June 30 in the journal Chemical Science. Feng Li and his collaborators also produced another research paper in the journal Analytical Chemistry, describing how to modify the nanomachine to detect diseases by examining a number of proteins in samples.
More information: Yongya Li et al. Simulation-guided engineering of an enzyme-powered three dimensional DNA nanomachine for discriminating single nucleotide variants, Chemical Science (2018). DOI: 10.1039/c8sc02761g
Journal information: Chemical Science , Analytical Chemistry
Provided by Brock University