Scientists reveal 'superbug's' artillery

Monash University's Biomedicine Discovery Institute (BDI) researchers have created the first high-resolution structure depicting a crucial part of the 'superbug' Pseudomonas aeruginosa, classified by the WHO as having the highest level threat to human health. The image identifies the 'nanomachine' used by the highly virulent bacteria to secrete toxins, pointing the way for drug design targeting this.
P. aeruginosa is one of a number of bacteria developing an alarming resistance to multiple drugs, raising concerns worldwide about the emergence of pan-resistant organisms.
Its virulence is due largely to the ability of the bacteria to secrete a suite of toxins and enzymes infecting the host environment.
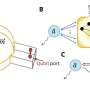
In a paper published this week in the online journal mBio, BDI researchers investigated a protein nanomachine on the surface of the bacterial cells responsible for the secretion of these toxins. The nanomachine, called the Type II secretion system, is responsible for the secretion of P. aureginosa's most toxic virulence factor, Exotoxin A.
"This is the first time we've seen how Pseudomonas aeruginosa secretes this important toxin," first author Dr Iain Hay said.
"This kind of first look is exciting and tells us that the next step of drug design may be feasible," he said.
"If you know the structure of this pore in the bacterial membrane that's pumping out the toxins that are important for virulence, you could design a molecular 'cork' to plug it."
Such a drug could potentially reduce virulence by stopping the secretion of toxins while other drugs worked at clearing the infection itself, Dr Hay said.
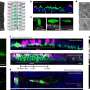
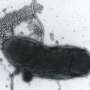
The researchers, led by Monash BDI's Professor Trevor Lithgow, used cutting-edge electron microscopy based at the Ramaciotti Centre for Cryo-Electron Microscopy (Monash University) to visualise the nanomachine pore. They used tens of thousands of images created by the microscope's beam to reconstruct a near-atomic resolution 3-D map of the 14-nanometre pore. A nanometre is a millionth of a millimetre.
"The Titan Krios microscope at Monash allowed us to see important molecular details of this nanomachine which have proved elusive for decades," Dr Hay said.
The methodology developed by the researchers would be applicable to other related bacteria surface nanomachines, he said.
More information: Iain D. Hay et al. Structural Basis of Type 2 Secretion System Engagement between the Inner and Outer Bacterial Membranes, mBio (2017). DOI: 10.1128/mBio.01344-17
Journal information: mBio
Provided by Monash University