Site-directed RNA editing as an alternative to genetic scissors
The development of CRISPR/Cas gene editing tools marked a revolution in the targeted alteration of genetic information. It opened up a wealth of opportunities for basic research and genetic repairs. Yet changing DNA has its risks – any errors it causes will be stored permanently in the genome and could therefore cause problems later on in both the individual and his/her subsequent offspring.
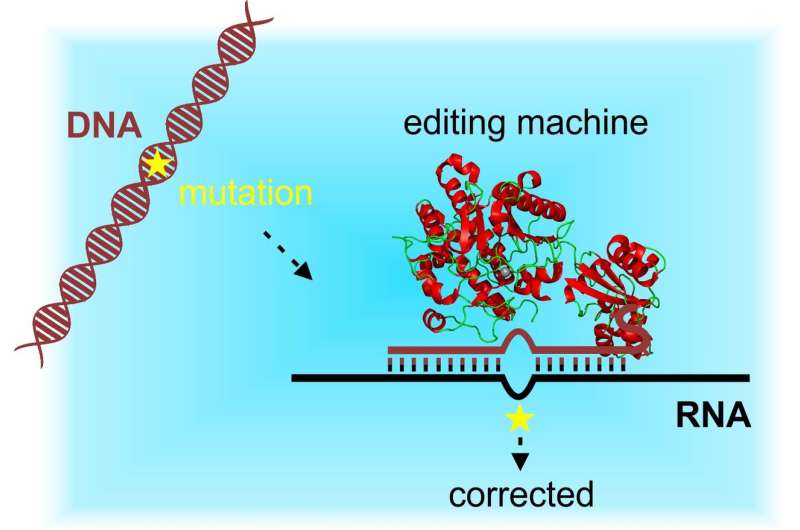
The research team headed by the University of Tübingen's Professor Thorsten Stafforst at the Interfaculty Institute of Biochemistry has been working for seven years on a low-risk alternative – seeking methods of targeted genetic alterations at the RNA level. The new method makes use of a normal cell process: the information encoded in DNA is forwarded by a working copy in form of RNA which is degraded when it is no longer needed. If one changes the RNA, the original message will remain in the DNA. Now the researchers have been able to precisely and efficiently edit these RNA transcript molecules inside the cell. The study has been published in the latest Nature Methods.
Cells copy the genetic information in their DNA – which is coded in a sequence of four different bases – into RNA molecules. The information becomes a set of instructions which is used to produce countless proteins in various compositions. The proteins in turn serve as building blocks in the cell and control its metabolism. "Our RNA editing methods are based on a protein construct which with the help of a guide RNA arrives at an RNA target molecule and converts certain bases. That makes it possible to rewrite the instructions for building proteins," Stafforst explains. RNA manipulation can be finely tuned and is reversible, making the method particularly attractive for treating diseases caused by mutations.
Time-limited manipulation
Stafforst points out that many disease-causing mutations might be accessible to the tool. He adds that the transient changes introduced in the RNA make it possible to intervene in signal pathways, e.g. associated with inflammation, whose permanent manipulation would have serious consequences. "We have already demonstrated the concurrent editing of multiple transcripts encoding signaling proteins," says Stafforst. The researchers showed that their method is twice as efficient as a recently published method for site-directed RNA editing, which is based on a variant of the CRISPR/Cas method.
Working together with the group of Professor Jin Billy Li at Stanford University, the Tübingen researchers demonstrated that the method makes only few mistakes – which the scientists were able to minimize. The method is highly efficient and many times more specific than competing methods, Stafforst added. The efforts, made for the application of site-directed RNA editing for basic research and medicine, are just the beginning. "In future, we want to omit engineered proteins and use naturally occurring enzymes instead to harness them for site-directed RNA editing," Stafforst says. The team has filed for a patent on their method.
More information: Paul Vogel et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs, Nature Methods (2018). DOI: 10.1038/s41592-018-0017-z
Journal information: Nature Methods
Provided by Universitaet Tübingen