Timing is everything: Researchers describe genetic clockwork in germ cell development
The nematode C. elegans is a true organizational talent: The tiny life forms, just one millimetre long, live for only two to three weeks, with sexual maturity lasting only four days. They nevertheless manage to generate over 300 offspring during this brief period. For this ambitious development programme to function optimally, a large number of processes must be synchronised within their cells. Geneticists at Martin Luther University Halle-Wittenberg (MLU) have deciphered a central signaling pathway that encodes and controls these processes. Their study was recently published in the Proceedings of the National Academy of Sciences (PNAS).
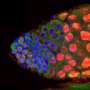
To reproduce, C. elegans must produce gametes, male sperm and female eggs. These develop from undifferentiated dividing stem cells. "Functional germ cells are special because they do not contain a double set of chromosomes and are no longer capable of division," says Professor Christian R. Eckmann, a developmental geneticist and Heisenberg Professor at MLU. Widely different programmes have to be sequenced in parallel to produce gametes: "On the one hand, the genetic material of the source cells must be halved in the cell division process. On the other hand, sexual differentiation into male and female has to take place, as well. Extensive intracellular restructuring is required to realize these processes, which have to mesh faultlessly if the cells are to develop successfully," Eckmann continues. A highly intricate clockwork mechanism with many interlocking gears gives some idea of the level of sequencing complexity involved.
These processes are controlled by RNA-binding proteins. Outside of the nucleus, in the cytoplasm, these proteins regulate selective gene activation. For a germ cell to develop out of a stem cell, two specific RNA-binding proteins need to be destroyed to reorganise the cell's genetic programme. How, when and why the signal for this developmental switch is given was previously unclear. The researchers from Halle have now figured out that the already familiar MAP kinase signalling pathway plays a central role. Eckmann summarises the process as follows: "A protein degradation cascade is initiated via this molecular pathway, at the end of which the two target proteins CPB-3 and GLD-1 are recognised, inactivated, and destroyed."
The geneticists at MLU were able to demonstrate that this process operates at a very early stage in meiosis and corresponds temporally to the sexual differentiation onset of female germ cells. The processes are thus optimally coordinated. According to Eckmann, "The special thing about these processes is that they involve known molecules with very long evolutionary histories, previously receiving attention as suppressors of tumour formation within the context of normal cell division. In C. elegans, these molecules were interleaved in an innovative way. The processes were adapted and temporally co-ordinated to facilitate optimized, rapid germ cell production." These findings of the MLU research group on Developmental Genetics suggest that the same genetic program may operate in germ cells of other, more complex organisms as well—albeit in a timely, less compressed form.
More information: Edyta Kisielnicka et al, MAPK signaling couples SCF-mediated degradation of translational regulators to oocyte meiotic progression, Proceedings of the National Academy of Sciences (2018). DOI: 10.1073/pnas.1715439115
Journal information: Proceedings of the National Academy of Sciences
Provided by Martin-Luther-Universität Halle-Wittenberg