Malaria's most wanted: Identifying the deadliest strains to design a childhood vaccine

Researchers have identified a 'genetic fingerprint' associated with the most deadly strains of malaria parasites, making these unique DNA regions potential targets for vaccine development.
An international research team led by the University of Melbourne found a small group of proteins was associated with the most severe strains of malarial infections, which are often fatal in young children who have not yet had a chance to develop a strong immune response to the parasite.
"We know that the great burden of mortality for malaria is in children under five," said Dr Michael Duffy, senior author on the study, from the University of Melbourne School of BioSciences and Bio21 Institute.
"But why children are at such high risk of death by malaria, and why some children die while others survive, has frustrated clinicians and scientists for years," he said.
"To better understand this difference, we compared the parasites causing the most severe malaria to parasites that cause uncomplicated or mild disease, which can be resolved by the immune system."
University of Melbourne Dean of Science Karen Day led a team that developed a 'fingerprinting' technique to identify different strains of malaria. The technique uses the parasite's var genes as a unique identifier. These genes code for different versions of the protein PfEMP1, which are expressed on the surface of red blood cells infected by malaria.
Each parasite has 60 of these genes that are different to other parasites and each gene is a mosaic of parts that can be shuffled to create new genes. The parasite also shuffles through the genes it uses like a pack of cards, thus appearing like different strains able to hide from our immune system.
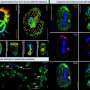
Dr Duffy and his colleagues used RNA sequencing to sample parasites isolated from the blood of 44 adults in a location in which malaria is endemic in the state of Papua, Indonesia. Twenty three of these people had severe malaria. The researchers then compared 4662 pieces of var genes that were being expressed in severe cases, against those expressed in mild cases.
"This is the first time anyone's taken the parasite genes that are expressed in the blood of patients and sequenced everything that's there. We then assembled these sequences to work out what's present and what's different between severe and uncomplicated cases," Dr Duffy said.
They showed that only 20 to 30 of the thousands of var genes were being expressed at a higher rate in patients with severe malaria than uncomplicated malaria. These 20-30 var gene pieces included many not previously investigated but also all of the proteins associated with severe malaria in India and Africa were also more highly expressed in the severe cases in Papua, suggesting that this small group of deadly proteins is highly conserved around the world, providing the promise of new targets for vaccine design.
The researchers are now looking to test children in malaria-endemic regions of Africa, the group by far the most at risk from malaria death, to see if the novel deadly proteins they found in Papua are also present there.
"We also want to look for serological responses - whether people have antibodies to these proteins in their blood" Dr Duffy said.
"Perhaps kids with severe infections are missing antibodies to these proteins, and those who don't get severe infections have them?"
A broadly effective malaria vaccine remains elusive despite the backing of the Bill and Melinda Gates Foundation and hundreds of millions of dollars in research funding.
Dr Duffy said that because malaria is so diverse, and each parasite is constantly changing its attack strategy, finding a valid target for a vaccine is hard.
"But now we are starting to understand that of the thousands of versions of the PfEMP1 protein that are out there, only a handful, maybe 20 or 30, are causing the most severe cases of malaria," he said.
"And, so, we might be able to target vaccines to just these severe versions.
"We won't eradicate malaria, but we may be able to protect children when they are most vulnerable to death or serious morbidity from malaria, and thus greatly reduce the burden of the disease."
The World Health Organization reported 429,000 malaria deaths in 2015, 70 per cent of those were children under the age of 5.
This research is published in PLOS Biology, and was conducted by an international team of researchers from the Universities of Melbourne and Oxford, the Walter and Eliza Hall Institute of Medical Research, the Eijkman Institute for Molecular Biology in Jakarta, Indonesia, the Timika Malaria Research Program, Papuan Health and Community Development Foundation, Indonesia, the Peter McCallum Cancer Centre and Charles Darwin University.
More information: Gerry Q. Tonkin-Hill et al, The Plasmodium falciparum transcriptome in severe malaria reveals altered expression of genes involved in important processes including surface antigen–encoding var genes, PLOS Biology (2018). DOI: 10.1371/journal.pbio.2004328
Journal information: PLoS Biology
Provided by University of Melbourne