Chromatin usage in individual cells reveals developmental trajectories
Both cell type and developmental stage can be deduced from measurements of chromatin accessibility in thousands of single cells, researchers at EMBL and the University of Washington show. They used this approach to uncover how cells in developing embryos regulate their identity as they decide what kind of cell to become. Nature publishes the results on March 14.
This new and more systematic approach allows researchers to analyse all the different cell types in an embryo at the same time, and importantly at a single cell resolution. "I expect this approach to save labs around the world lots of time," says Eileen Furlong, who co-led the work at EMBL in Heidelberg with Jay Shendure at the University of Washington School of Medicine in Seattle.
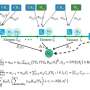
Previously, a researcher would have to first isolate the various cell types, and then investigate the chromatin of each type in separate batches. That lengthy method provided an averaged view across thousands of cells of a given cell type. "Previous studies have used differences in RNA content to identify cell types and their developmental trajectories," says Shendure. "Here, we instead measure the state of chromatin in single cells, which contains the regulatory program that governs how and when RNAs are expressed in each cell."
"For the first time, we have looked upstream at how these expression signatures are regulated and therefore drive single cell trajectories during early development," Furlong adds.
Role of chromatin
Chromatin is the tightly-coiled structure of DNA and proteins which is used to store the genetic information inside the nucleus of a cell. The chromatin in a human cell contains around two metres of DNA, packed into a nucleus less than one hundredth of a millimetre across. Regulatory elements like promoters and enhancers are short stretches of DNA that regulate the levels of gene expression and therefore the production of proteins, which are what ultimately make cell types different from one another. When cells use a particular regulatory element, the chromatin uncoils and its content becomes accessible. That's why Furlong, Shendure and colleagues expected chromatin accessibility to shed light on how a cell follows a specific developmental path, turning into a highly specialised muscle or nerve cell, for example.
Having single-cell information on chromatin accessibility allowed the team to determine a cell's identity and how it is regulated. They performed the experiments on fruit fly embryos, a very important model organism for both developmental biology and disease models, but the approach can be applied to any species. The results identified thousands of previously unknown regulatory elements that are used only in a subset of cells and predicted when and where each element are active during development. The data, made available through a user-friendly browser (see below), reveals a wealth of differences between cell types and provides a powerful resource for future studies.
This work was the result of a collaboration between James Reddington and David Garfield in the Furlong group at EMBL, and Darren Cusanovich in Jay Shendure's group at the University of Washington School of Medicine, Seattle. Looking forward, the research team plans to expand this study, and to integrate other layers of single-cell information on the regulation of cell fate decisions during embryogenesis.
More information: The cis-regulatory dynamics of embryonic development at single-cell resolution, Nature (2018). nature.com/articles/doi:10.1038/nature25981
Journal information: Nature
Provided by European Molecular Biology Laboratory