Enzymes and bacteria move directionally, but bacteria towards food, enzymes away
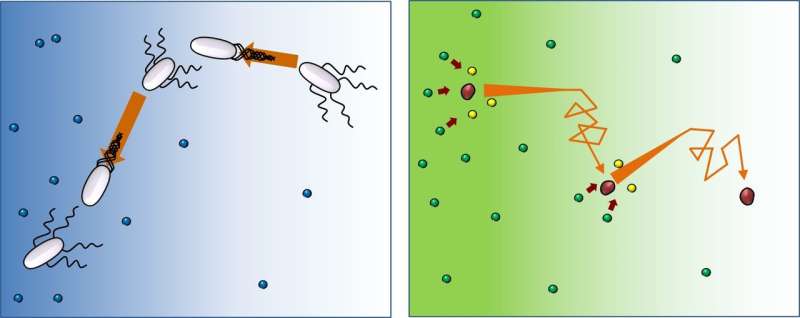
Although putting together the words "random" and "biased" can seem like a clash, these are the attributes that describe how bacteria navigate and fetch their food. They follow a random path, but biased toward the direction of the nutrients' source. Scientists at the Center for Soft and Living Matter, within the Institute for Basic Science (IBS, South Korea), observed that enzymes, the molecules that help biological reactions to happen more quickly, move with a similar pattern, but in the opposite route—randomly traveling towards the area with fewer substrates. These results, obtained with new optical technologies and published as an inaugural article in Proceedings of the National Academy of Sciences (PNAS), are particularly intriguing as they comprise a departure from the traditional view.
Thousands of enzymes promote a multitude of reactions in our bodies, each of which sustain our lives. Enzymes act upon other chemicals called substrates and speed up chemical reactions that convert substrates to products. To do that, enzymes and substrates have to meet. It was originally believed that enzymes and their correspondent substrates casually bump into each other: "Standard textbooks mention that enzymes and substrates are supposed to come close 'somehow," interact, and that's how reactions happen to take place. We tried to dig into the 'somehow' part of this process. After collecting thousands of events we concluded that enzymes' walk is random biased, meaning that they are microscopically directional, like bacteria," explains Ah-Young Jee, first author of this study.
The motion of swimming bacteria is known as "run-and-tumble": run, turn and repeat. They proceed in one direction, then change direction randomly, and repeat. However, while bacteria orient toward the food supply, enzymes move in the direction of lesser substrate concentration. "Molecules lack decision making capabilities, but surprisingly move toward areas with less substrate. Indeed, the substrate introduces the direction bias, and the higher the substrate concentration we used in the experiment, the stronger this tendency. We hypothesize that it could be a way to make the product concentration uniform around the medium, even when substrate concentration is not," says Prof. Tsvi Tlusty, who offered the theoretical understanding of this research.
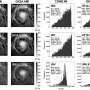
As each enzymes' stride is only around 50 nanometers in length, extremely precise devices are needed to measure their speed and orientation. The research team, led by Steve Granick, looked at the enzymatic motion with a super-resolution microscopy technology, known as stimulated emission-depletion fluorescence correlation spectroscopy, or STED-FCS for short.
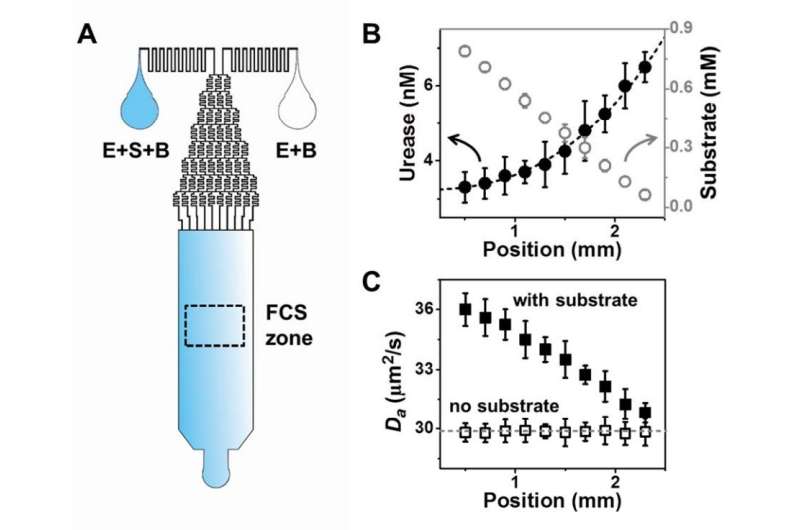
In the FCS method, enzymes are decorated with fluorescent molecules and their passage through a small dot is detected by a laser beam. Statistical information is gathered, such as the number of enzymes passing through the dot and their speed (diffusion rate). The team complemented FCS with STED technology, one of the strengths of this research center. STED concentrates the laser beam into an extremely small area, allowing a more refined measurement of the enzymes' position. The FCS laser beam covers an area of around 250 nanometers in diameter, which is approximately 25 times the diameter of the enzyme (10 nanometers). STED-FCS is more accurate since it reduces the beam diameter to 50 nanometers, which is comparable to the distance of an enzyme's step.
The researchers also designed a new microfluidics chip, formed by micrometer-thick channels where liquids can pass through. Previous studies, using FCS without STED and a different microfluidics chip, suggested that the enzymes move towards the area with the highest concentration of substrate. However, this new technology allows deeper observations and showed the opposite. The authors think that the results differ because detailed information about the enzyme motion was canceled out using a larger laser beam. It would be similar to localizing someone within 50 square kilometers or within 2 square kilometers, the latter is more accurate. They created conditions where the enzyme is uniformly distributed inside the chip, but the substrate concentration varies from left to right: from a minimum concentration on the right side of the chip, to a maximum concentration on the left. Then, they used FCS to observe the enzyme's concentration and speed inside the chip, and found that the enzyme tends to accelerate towards the area with lesser substrate concentration (that is towards the right side of the chip). Instead, when no substrate is present, the enzymes do not change their speed. Moreover, the super-resolution capabilities of STED-FCS showed run-and-tumble dynamics.
The team observed the same behavior with two enzyme-substrate couples separately: urease-urea and acetylcholinesterase (AChE)-acetylcholine. The former enzyme is used by some microorganisms to change urea into ammonia and carbonic acid, the latter plays an essential role in neuromuscular junctions.
In the future, the research team aims to expand the complexity of the study and test conditions more similar to real life: "This study looked at the behavior of one enzyme in response to one substrate. Next, we plan to look at several enzymes at the same time. Our body functions with a cascade of reactions, meaning that one reaction triggers the next: enzyme A triggers enzyme B, then B triggers C, and so on. STED-FCS analysis could be a key tool to advance metabolic research by revealing how these multiple enzymatic systems are organically related," concludes Jee.
More information: Ah-Young Jee et al, Enzyme leaps fuel antichemotaxis, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1717844115
Journal information: Proceedings of the National Academy of Sciences
Provided by Institute for Basic Science