Establishing a genome-wide map of bacterial genes crucial for colonization of plants by beneficial microbes
Working with the plant growth-promoting bacterium Pseudomonas simiae, researchers have identified 115 genes that negatively affect its ability to colonize a plant root system when mutated.
A plant's health and development is influenced by the complex community of microbes that surround it. By identifying the bacterial genes that can alter how well microbes can colonize a plant, researchers can develop targeted approaches to improve plant health and growth for a number of applications, including increased biomass yield for biofuel production.
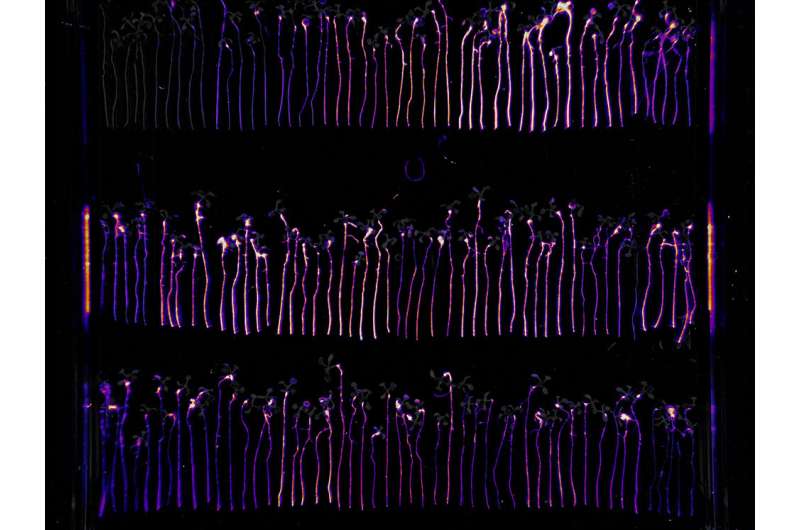
A plant's health and development is influenced by microbes residing within the plant (endophytes), in the soil, and in the narrow region where the plant roots interact with the soil (rhizosphere). To better understand how microbes colonize the root environment, researchers at the Joint Genome Institute, a DOE Office of Science User Facility, and their collaborators at the Howard Hughes Medical Institute at the University of North Carolina, applied a genome-wide transposon mutagenesis approach on the model plant growth-promoting bacterium Pseudomonas simiae using the model plant Arabidopsis thaliana as a host to generate a genome-wide map of bacterial genes that affect the efficacy of microbial colonization.
Through randomly barcoded transposon sequencing (RB-TnSeq), the team identified 115 genes that, when mutated, have reduced root colonization capabilities. These genes are involved in functions such as sugar metabolism, cell wall synthesis, and motility. The team also identified 243 genes that, when mutated, positively alter root colonization capabilities, many of them likely involved in amino acid transport and metabolism. Additionally, the team identified 43 genes to which very little or no functional information could be assigned. The researchers suggested that these genes may represent novel functions or pathways yet to be characterized. The work shows that RB-TnSeq can be applied to assess in vivo bacterial plant root colonization.
Among the contributors to this project was Sabah Ul-Hasan, a 2015 intern through the DOE JGI/University of California, Merced Genomics Distinguished Graduate Internship Program. The program offers UC Merced graduate students hands-on experience in cutting-edge genome research as part of the DOE JGI's commitment to training the next generation of scientific talent.
One of the principal challenges arising from rapid sequencing is the assignment of functions to new genes. The RB-TnSeq approach used here can accelerate the association of new genes with characteristics and behaviors of importance to DOE missions such as understanding how microbes help (or hinder) the growth of crops that could serve as bioenergy feedstocks. Getting at the fundamental genetic contributors, such as the 115 genes that negatively regulate microbe-plant root interactions, will help focus future efforts to advance this research.
More information: Benjamin J. Cole et al. Genome-wide identification of bacterial plant colonization genes, PLOS Biology (2017). DOI: 10.1371/journal.pbio.2002860
Journal information: PLoS Biology
Provided by DOE/Joint Genome Institute