Experiments with bacteria show that genes can fuse together, leading to the production of novel proteins
All organisms must continuously adapt to their environment in order to survive. Such adaptation is brought about by changes in their genetic material. Together with colleagues from New Zealand, Paul Rainey from the Max Planck Institute for Evolutionary Biology in Plön has been studying the emergence of new, better adapted cell types in the laboratory. The researchers have discovered that one mechanism by which bacteria can develop new characteristics is through the fusion of two existing genes. In some of the cells, this resulted in genes coming under the control of a new promoter, resulting in the synthesis of larger quantities of the protein encoded by the gene. In another case, two neighbouring genes fused together. The protein encoded by the resulting gene – composed of parts of the two original genes – has a different localization within the cell. This effect is also known from other organisms, including humans. A gene fusion of this type results in bacterial cells which are better adapted to their environment.
Changes to the genetic code in existing genes – mutations – can equip an organism with new characteristics. Duplication of genes and the insertion of extra sections of DNA can also increase an organism's adaptability. Over the course of evolution, it is even possible for completely new genes to be created. This involves changes in previously non-functional stretches of DNA, which result in them becoming functional templates for protein synthesis. Another known mechanism of gene creation is the fusion of two genes, resulting in the production of a novel protein.
"These insights have been obtained by comparing the genomes of different organisms. Because evolution usually works very slowly, it is not usually possible to observe this kind of change in genetic material in real time – let alone how it affects the survival of the organism in question," says Paul Rainey of the Max Planck Institute for Evolutionary Biology. The researchers therefore focused on bacteria, which reproduce extremely quickly and can be bred in the laboratory in huge numbers in a small space. This allows researchers to study the resulting changes in the genome and actually observe evolution in action.
Survival in a lab environment
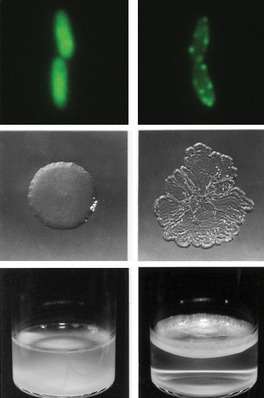
Rainey's laboratory experiments look at how the bacterium Pseudomonas fluorescens evolves characteristics relevant to survival in liquid-filled laboratory microcosms. The ancestral type grows in the broth phase, but depletes essential oxygen setting the scene for mutants that form mats that access oxygen at the air-liquid interface. Mutants that form mats are easily detected because of their unique wrinkled colony morphology.
According to Rainey and his colleagues, mat formation is caused by a variety of mutational changes in genes that regulate di-guanylate cyclase activity. These mutations switch off negative regulators causing the di-guanylate cyclases to assume an active state. When the researchers eliminated pathways subject to negative regulation they discovered a set of previously unknown mutations that caused the wrinkly mat-forming phenotype. In some of these, the di-guanylate cyclase gene had come under the control of a different promoter, resulting in increased di-guanylate cyclase production.
Fusion protein relocalizes
In some wrinkly cells, however, the activity of this gene was unchanged. Analysis of the mutations in these cells showed that these mutants contained a chimeric gene formed from the di-guanylate cyclase gene and a neighbouring gene. The protein encoded by the latter is normally active in the cell membrane. "There must, therefore, have been a fusion event between two genes encoding proteins which are usually found at different locations within the cell," explains Rainey. The new protein possesses a membrane domain and is embedded in the cell membrane. This activates the protein, resulting in increased cellulose production.
In other organisms too, proteins produced by gene fusions frequently end up having a different localization within the cell. In humans, for example, the Kua-UEV gene is the result of the fusion of the Kua and UEV genes. The new UEV protein now localizes to internal cell membranes and performs a new function. In humans, 64 percent of gene families for mitochondrial proteins contain a gene for a protein which is active elsewhere in the cell. "Although in our experiments gene fusion events made up only about 0.1 percent of mutations resulting in the wrinkly phenotype, outside the laboratory they may be more common," says Rainey.
More information: Andrew D. Farr et al. Adaptive evolution by spontaneous domain fusion and protein relocalization, Nature Ecology & Evolution (2017). DOI: 10.1038/s41559-017-0283-7
Journal information: Evolutionary Biology , Nature Ecology & Evolution
Provided by Max Planck Society