Researchers use new techniques to pinpoint evolution in fungi
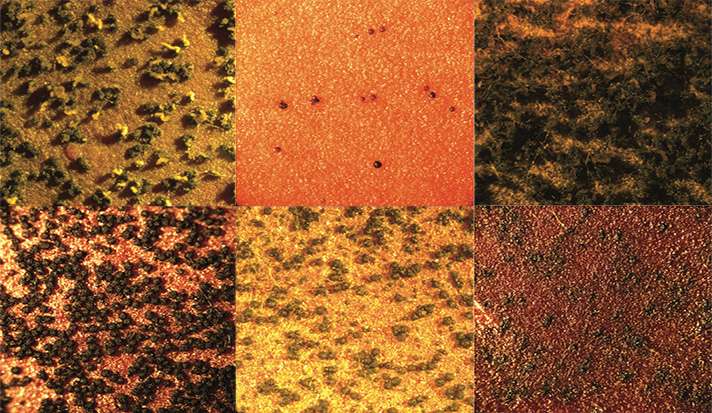
Authors from Yale and Michigan State University collaborated on a National Science Foundation study of five types of fungi that has illuminated a successful new strategy for pinpointing genes responsible for the evolution of certain biological processes.
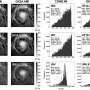
To better understand the effects of genes on the evolution of an organism, large teams of biologists previously have worked to "knock out"—or turn off—thousands to tens of thousands of genes within the genome, seeing if they can perceive an effect that the gene knock-out has on the organism. However, this new work demonstrates a more precise shortcut for researchers: If they first identify identical genes present across different species that have increased in gene expression during their recent evolutionary development, the scientists can "knock out" this smaller targeted set to reveal genes underlying an organism's phenotype.
This is exactly what Jeffrey Townsend of Yale, and Frances Trail of Michigan State did, discovering many genes that play roles in the development of fungal fruiting bodies. "We've now demonstrated—for the first time—that you can predict genes that relate to novel phenotypes by estimating which genes have increased their expression recently in evolutionary time," wrote Townsend.
This discovery has important implications for efficiently discovering the genetic basis of evolved traits, including desirable traits in agriculture and animal husbandry and undesirable traits in invasive species and in plant, animal, and human pathogens, say the researchers.
More information: Frances Trail et al. The ancestral levels of transcription and the evolution of sexual phenotypes in filamentous fungi, PLOS Genetics (2017). DOI: 10.1371/journal.pgen.1006867
Journal information: PLoS Genetics
Provided by Yale University