Revealing the structure of an elusive quality control enzyme

The structure of the critical quality control checkpoint enzyme that oversees the production of thousands of secreted glycoproteins has been solved by a fruitful collaborative effort at Diamond Light Source. The study, recently published in PNAS, found that the enzyme had surprising flexibility that allowed it to adapt its conformation and clasp its client glycoproteins.
Glycoproteins are an abundant type of protein that have sugars known as glycans attached to them. To ensure that glycoproteins are correctly folded, they must be scrutinised by a quality-control enzyme known as UDP-glucose:glycoprotein glucosyltransferase (UGGT). Incredibly, the enzyme has the capability to check and detect misfold in thousands of proteins of all different shapes and sizes, but the mechanism of this impressive feat has yet to be revealed. This important enzyme has been studied for the past 25 years, but its structure has eluded all who have worked on it thus far.
Drawn to the challenge, a concerted effort was made by academics at the University of Oxford and the National Research Council of Italy, along with staff at Diamond, to finally determine the structure and solve the mystery of this enigmatic enzyme. The structure was solved with the aid of the Macromolecular Crystallography beamline (I04-1) and cryo-electron microscopy (EM) at the state-of-the-art Electron Bio-Imaging Centre (eBIC) both at Diamond.
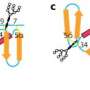
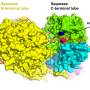
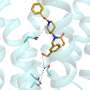
The team saw that UGGT had seven subunits rather than the four that were expected from sequence, and that it was very flexible. These qualities would allow the enzyme to be highly promiscuous, as it could adapt its conformation to fit the proteins it checks. These fascinating discoveries could facilitate the design of novel UGGT inhibitors that might impair the folding of viruses to treat infections or could release actve and yet retained proteins to treat rare congenital diseases.
Elusive enzyme
Glycoproteins make up a huge proportion of the protein content of cells. The majority of secreted proteins are glycosylated and even viruses hijack this pathway to be folded correctly to spread their infection. The critical governor of the folding quality of glycoproteins is UGGT, a 170 kDa enzyme that is found in all eukaryotes, from yeast to fish to birds and mammals. UGGT acts as the gatekeeper for glycoproteins by flagging any that are misfolded and preventing their premature release from the endoplasmic reticulum. Although UGGT is widespread, its structure and function has eluded scientists for 25 years. Its intriguing promiscuity for checking thousands of glycoproteins of different shapes and sizes has attracted much attention.
Scientists from the University of Oxford, the Institute of Sciences of Food Production and the Institute of Crystallography at the National Research Council, Italy, along with a team from the eBIC at Diamond embarked on a groundbreaking structural study to delve into the inner workings of UGGT.
Lead scientist of the joint effort and research scientist at University of Oxford, Dr Pietro Roversi, explained their motivation: "We wanted to know how UGGT could be in charge of checking the correctness of folded proteins given that they are all so different. There are some very important targets of UGGT, including immunological proteins and those that are retained in rare congenital diseases."
Heat-resistant UGGT
One of the reasons why the structure of UGGT had eluded scientists for so long was because of its flexibility. To overcome this hurdle, the team wisely chose to study a form of UGGT derived from a thermophilic fungus. Proteins from heat-resistant sources can often be more rigid, meaning that this type of UGGT was less flexible and more amenable to structural analyses than its human counterpart.
While the crystal structure was solved by Dr Roversi at I04-1, an expert team from Diamond worked concurrently at the eBIC to solve the cryo-EM structure.
Principal investigator of the study and Professor of Virology at the University of Oxford, Nicole Zitzmann explained their findings: "We saw that UGGT was made up of more domains than anticipated, which could not have been predicted from the sequence alone. There were seven domains in total: a catalytic domain, two β-sandwiches and four thioredoxin-like domains." One of the biggest discoveries was the high flexibility of UGGT, which if impaired prevented the enzyme from functioning. It is this flexibility that allows it to clasp and adapt its shape to check its vast number of client proteins.
UGGT inhibition
As well as furthering our basic knowledge of how this important quality checking protein works, the study could give rise to novel UGGT inhibitors. It is hoped that antagonising UGGT could enable viral infections or rare congenital protein storage disorders to be treated. A further important application could be to improve protein expression systems in eukaryotic cells, whereby loosening the control that UGGT exerts could increase yields of secreted proteins.
Dr Roversi described the next steps for the study: "We want to solve the structure in complex with misfolded client glycoproteins, but we also want to carry out basic cellular biology to see which pathologic glycoproteins proteins UGGT has the power to retain in the endoplasmic reticulum, so that we can ascertain in which diseases this enzyme is implicated."
More information: Roversi P et al. Inter-domain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint. PNAS, (2017) DOI: 10.1073/pnas.1703682114.
Journal information: Proceedings of the National Academy of Sciences
Provided by Diamond Light Source