Scientists investigate how the sense of smell works in bacteria
Scientists from MIPT, in collaboration with international colleagues, have proposed a universal mechanism for the sense of smell in bacteria. The researchers obtained the structure of the NarQ protein from Escherichia coli (E. coli), which belongs to a universal class of sensory histidine kinases that are responsible for transmitting signals to bacteria about their environment. The paper published in Science describes how bacteria communicate with one another and form biofilms on sterile surfaces or inside the human body.
Drugs affecting this sense of smell could potentially be used as substitutes for modern antibiotics. They do not kill bacteria; they simply present signals that render them harmless to the human body. Theoretically, bacteria cannot develop resistance such an attack .
The two components of a cell's sense of smell
All cells are encased by a dense membrane through which virtually no chemicals can pass. This enables the cell to keep its internal chemical conditions constant and functioning correctly. However, the membrane greatly limits the exchange of information with the environment. In order to sense what is happening on the outside, a cell uses special molecular machines called a proteins. These proteins very often exist in the membrane itself or close to it, and they are responsible for transmitting signals or chemicals into or out of the cell.
The most universal mechanism for bacteria to "sense" an environment are so-called two-component systems. Such systems consist of two proteins: a kinase, which receives the signal from outside of the cell and transmits it into the cell, and a response regulator, which receives the signal inside the cell and triggers subsequent reactions.
Molecular photography
A useful method of understanding how proteins function is to observe their structure at the atomic level. At present, most protein structures (more than 100,000) have been obtained using X-ray crystallography. This method involves observing the diffraction pattern of protein molecules ordered in a crystal lattice pattern. However, this only reveals the structure of one state of the protein, as in a photograph. If researchers can image the initial and final state of a process, they can make a guess as to how, exactly, the protein works when switching between these states.
Membrane "pistons" power a cell's sense of smell
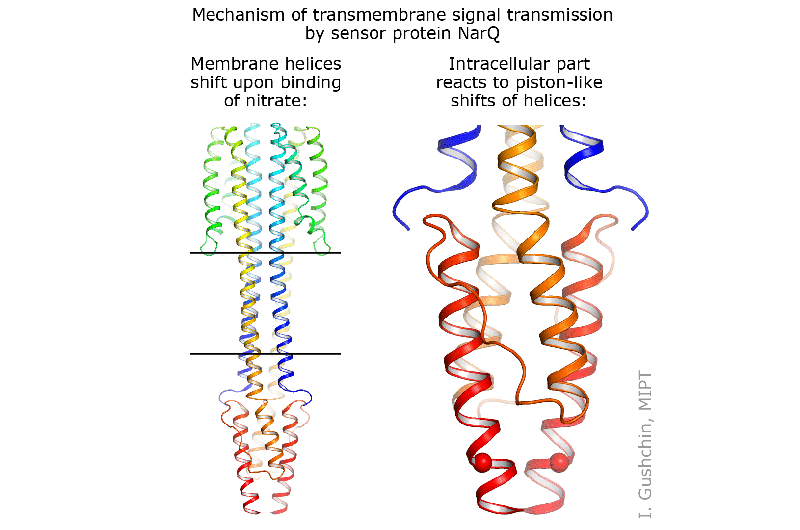
The authors of the study were able to obtain the structure of two states of the NarQ kinase from E. coli. This kinase senses the presence of nitrates in the environment and sends a corresponding signal through the cell membrane. As it turns out, the sensor in both of its two states is a dimer, two protein molecules working together to capture the nitrate. The first state is inactive—the protein is not bound to the nitrate ion and does not transmit a signal. The second state is active, or signaling—in this state, the kinase transmits a signal into the cell to inform it that nitrates are present in the environment.
The protein structure in the active state was obtained for the most reliable wild-type protein, one without artificial mutations, which scientists often use to increase the stability of a protein. To obtain the structure in the inactive state, the authors mutated the site that the nitrate binds to. The stability of the protein was not affected; however, the nitrate no longer became bound to it, which gave the authors the opportunity to observe a kinase in a state of inactivity.
It was found that the signaling and inactive states differ only very slightly at the nitrate-binding site, by 0.5-1 angstroms, which is approximately one-fifth of the size of the ion itself (1 angstrom is 10-10 meters). However, when this ion binds to the sensor, it causes huge changes in the protein. The helices of different monomers begin to move in different directions, like pistons. Such molecular pistons transmit the small change of 0.5-1 angstroms through the membrane, and their outer ends shift by approximately 2.5 angstroms in different directions. Inside the cell, in the HAMP domain, these shifts are converted into the rotation of two parts of NarQ relative to each other. Ultimately, the positions of the output helices change by as much as seven angstroms, thus completing the signal transmission.
Aside from the structures in which the two proteins form a symmetrical pair, the scientists were able to produce a structure with an asymmetric position of the two proteins. In this state, the protein is arranged differently in the crystal and is bent strongly. However, the effect on the regulator remains virtually unchanged. This versatility of observed movement reveals that the signal transmission mechanism is universal, and that sensors of other chemical compounds operate via the same piston-shift mechanism.
"How signals are transmitted through the cell membrane is one of the most fundamental questions in modern biology. In this study, we showed in detail how a signal (in this case the binding of a nitrate) can be transmitted by hundreds of angstroms into the cells of bacteria and archaea, as well as fungi and plants. With a better understanding of the mechanisms of signal transmission, we can expect to learn how to manipulate such cells, and in particular, to try to weaken or neutralize the harmful effects of pathogenic microorganisms," said Ivan Gushchin, the head of MIPT's Laboratory of Structural Analysis and Engineering of Membrane Systems, commenting on the study.
More information: Ivan Gushchin et al, Mechanism of transmembrane signaling by sensor histidine kinases, Science (2017). DOI: 10.1126/science.aah6345
Journal information: Science
Provided by Moscow Institute of Physics and Technology