New protein discovery may lead to new, natural antibiotics
Scientists have discovered a new protein that likely will advance the search for new natural antibiotics, according to a study by Texas A&M AgriLife Research published Feb. 13 in the journal Nature Microbiology.
The discovery has to do with how proteins regulate gene expression. Scientists know a great deal about proteins that control how certain gene clusters get their start—referred to as transcription initiation—but much less is known about transcription elongation where proteins keep gene expression going through "roadblocks" in the DNA sequence, according to Dr. Paul Straight, AgriLife Research biochemist in College Station and the paper's co-author.
"The upshot is that our discovery expands the basic knowledge of processive antitermination—a type of genetic regulation—and demonstrates that the mechanism is more widespread among bacteria than previously thought," Straight said. "Antibiotic production by bacteria involves complex chemistry that is often encoded in a collection or 'cluster' of many genes. To express these giant gene clusters requires special regulation mechanisms. Understanding these mechanisms could help a great deal in the search for new antibiotics produced by bacteria."
For this study, Straight and his graduate assistant Chengxi Zhang of College Station, teamed up with University of Maryland researchers Jonathan R. Goodson, Steven Klupt and Dr. Wade Winkler.
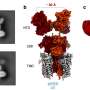
The team describes two discoveries in the paper. One is the protein they named LoaP, which stands for long operon associated protein. The other discovery is that the protein is frequently found next to the gene clusters that are responsible for producing antibiotics. Hence, knowing how LoaP works and its prime location could lead scientists to a shortcut for antibiotic production.
Straight explained that bacteria frequently string together many genes, which are expressed as a group when the cell needs the proteins the genes encode.
"These long chains of genes raise challenges for the molecular machines that decode DNA. Sometimes the molecular machines hit roadblocks, called terminators, and they stop and fall off the DNA. The LoaP protein is called a processive antiterminator, because it helps the machines stay on DNA and move through the roadblock terminators."
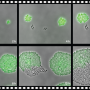
The team discovered the LoaP protein in Bacillus amyloliquefaciens, a bacteria known to ward off pathogens that attack plant roots in agriculture, aquaculture and hydroponic production.
The researchers said the Earth has an abundant and diverse supply of microbes such as bacteria and many of them have useful biomedical purposes. While researchers realize the potential benefits—and indeed have been studying them for more than 100 years—a modern limitation to antibiotic discovery is the murky understanding of the genetic regulatory mechanisms that oversee their production.
"After nearly a century of searching for bioactive natural products, bacteria still constitute a major target of modern drug discovery," Straight said. "The characterization of the biochemical pathways of these molecules remains a bottleneck to their development. One of the key restrictions is a shortage of knowledge on the range of genetic mechanisms that can affect them. Therefore, the discovery of new classes of genetic regulatory mechanisms is likely to impact future development of natural products that counter disease."
More information: Jonathan R. Goodson et al, LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens, Nature Microbiology (2017). DOI: 10.1038/nmicrobiol.2017.3
Journal information: Nature Microbiology
Provided by Texas A&M University