Watching the ribosome at work
A new statistical method could help to clarify the function of unknown genes. A research team under Uwe Ohler of the Berlin Institute for Medical Systems Biology (BIMSB) at the Max Delbrück Center for Molecular Medicine (MDC) has adapted and tested a filter method from speech signal processing that makes sequencing data more interpretable. "We can use this to watch the ribosome at work," says Ohler.
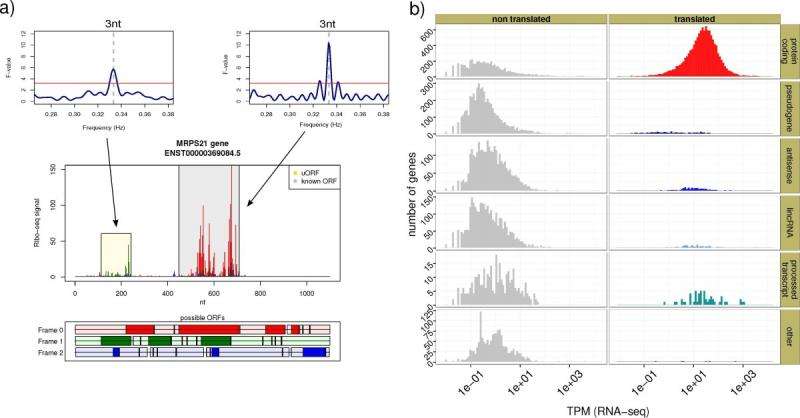
Ohler, a computer scientist, focused on speech recognition programs during his studies. He made use of statistical procedures to filter out relevant information from the background "noise pollution" surrounding the data and thus identify words accurately. The mathematical methods used for this purpose, which include the Fourier transform, have been indispensable to modern data processing for some time. Astrophysicists investigating spectra in the light from distant stars or developers working on speech recognition for mobile phones face the same challenge: "noisy" signals need to be interpreted as accurately as possible. Now Ohler is applying these filtering methods to molecular biology. With his colleagues from the Berlin Institute for Medical Systems Biology (BIMSB) at the MDC he has developed and tested RiboTaper. The program filters the relevant information out of certain sequencing data to determine whether one of the cellular protein factories - ribosomes - is actually active on the RNA.
RiboTaper is based on a laboratory procedure that was developed several years ago in the United States. It is called Ribo-seq and is used to identify the part of a gene that encodes a protein. This is important, as the theory that all genes encoded in the DNA contain a "construction manual" for a protein is not entirely accurate. Thousands of genes that have been mapped in the genome in recent years are indeed transcribed in the RNA, but it is not known whether they contain small, protein-coding sections. Overall, only a small part of the genome is responsible for producing proteins. The lion's share of the DNA has regulatory functions. Furthermore, from cell to cell, different genes are sometimes up and sometimes down regulated, or are shut down. How can we find out which genes in which cells actually produce protein and which do not?
The answer can be found by looking at the ribosomes and the construction manual that they work from. Ribo-seq helps with this, because this wet lab procedure in effect "freezes" the ribosomes in their place on the RNA strand. The RNA is the construction manual transmitted from the genes. Everything except ribosome and the associated RNA are digested using biochemical tools. This allows the molecular biologists to determine which instructions the ribosomes are working with. The problem is that the data obtained with Ribo-seq is "noisy." There are tiny remnants of DNA, RNA and proteins that occur naturally and are dismantled in each cell. Furthermore, one never knows exactly whether the ribosomes are really active, and produce proteins, at the identified point on the RNAs, or whether they are, in effect, just waiting for another signal. The dry lab method RiboTaper should help to fill this information gap. It can be used to clarify the roles of DNA, RNA and ribosomes much more precisely.
"We know, for example, that a specific ribosome usually covers some 29 RNA building blocks, or nucleotides," says Ohler. "And we also know that the ribosome moves along the RNA in intervals of three nucleotides." This creates a periodic pattern, which bioinformaticians can search for in all the data. "This then shows us the points on the RNA where something significant is happening," says Ohler. You can begin to imagine what this is like if you think of a kitchen that has been gutted by fire. Forensics investigate the kitchen and find recipe sheets, sugar, eggs and flour. But was the cake ready when the kitchen was on fire? Or were only the ingredients for the dough ready? What did the cook intend to bake? Using Ribo-seq in combination with RiboTaper, molecular biology forensics is much closer to finding out the secret of the cellular kitchen.
Ohler explains: "With RiboTaper we can hunt down smaller proteins in previously poorly studied genes and help to clear up conflicting data interpretations." Ohler also sees another advantage: "Sequencing devices are now available in many laboratories, but only a few centers also have access to a good mass spectrometer. With RiboTaper we can draw conclusions about which transcripts are actively translated into proteins." To test the new procedure, Ohler put his samples to the test and had the RiboTaper data checked by his MDC colleague Matthias Selbach, using mass spectrometry. Since a number of groups at the MDC are already using Ribo-seq, RiboTaper may be able to assist them in their interpretation in exciting new ways.
Ohler's laboratory collaborated with colleagues from the BIMSB on this study, including research groups led by Markus Landthaler, Benedikt Obermayer and Matthias Selbach.
More information: Lorenzo Calviello et al. Detecting actively translated open reading frames in ribosome profiling data, Nature Methods (2015). DOI: 10.1038/nmeth.3688
Journal information: Nature Methods
Provided by Helmholtz Association of German Research Centres