June 10, 2015 report
Blood test for cancer biomarkers using an electrochemical clamp assay
(Phys.org)—Researchers have found an innovative way to detect cancer biomarkers in a person's blood. Nucleic acids, the components of DNA and RNA, are typically located within the cell. However, sometimes these nucleic acids can be found circulating in the blood. Cancer patients tend to have more of these cell-free nucleic acids in their blood. A small portion of these cell-free nucleic can contain mutations associated with certain cancers.
Most methods of studying cell-free nucleotides involve DNA sequencing or PCR studies. Both of these have drawbacks. DNA sequencing is expensive and patients often have to wait several weeks for results. PCR requires extensive sample preparation and modifications to make it sufficiently selective for point mutations. To improve upon these techniques, Jagotamoy Das, Ivaylo Ivanov, Laura Montermini, Janusz Rak, Edward H. Sargent, and Shana O. Kelley from the University of Toronto and Montreal Children's Hospital have developed a method that selectively identifies mutations common in lung and skin cancer with little-to-no sample preparation. Their work appears in Nature Chemistry.
Over the past two decades, cancer genomics has made great strides in identifying genetic markers for certain cancers. Two examples used in the current study are the KRAS and the BRAF markers. KRAS has several mutations associated with it that are indicators of lung, colorectal, and ovarian cancer. BRAF mutations are most notably associated with melanoma, the most deadly form of skin cancer.
Many genetic cancer markers involve a point mutation on a particular gene. Point mutations are difficult to detect because they are in such small quantities in blood compared to normal cell-free nucleic acids, or wild type. One way that scientists have made PCR more sensitive is by using genetic "clamps" called Peptide Nucleic Acids (PNAs). These are strands of a complementary nucleotide sequence that bind to wild type sequences and consequently, amplifying the target sequence.
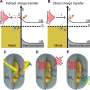
Das, et al. combined this PNA clamp technique with an electrochemical probe to make a fast, selective, and cost-effective sensor. The first designed a clamp system to select for particular mutations in the KRAS gene. KRAS has seven mutations that are associated with lung cancer. Their model system included clamps for all of the other possible mutations except for 134A, their target mutation, and it had clamps form wild type sequences.
They designed an electrochemical probe functionalized with complementary nucleic acid strands to various target sequences. The functionalized probe is a nanostructured microelectrode made from using nanolithography to insert palladium-coated gold deposits on silicon-coated wafers. The nanostructured microelectrodes were functionalized with PNAs that were specific to the 134A mutation sequence. Once the target sequence binds to the probe, it is read using an electrocatalytic reporter pair (Ru(NH3)63+ and Fe(CN)63-). Based on the electrochemical signal, their probe selectively bound the 134A mutation sequence without binding wild type sequences.
To optimize their clamp assay, Das et al. tested whether their system could detect all seven mutations in KRAS in a non-purified, complex mixture of complementary mutant target, non-complementary mutants, the wild-type sequence, total human RNA and the clamp cocktail. They found that all seven mutations can be selectively detected using their system based on which PNA is omitted from the mixture.
The next step was to see if the wild type PNA clamp needed to be included in the system for selective detection of point mutations. They tested a mutant positive patient sample and a healthy donor sample, and tested with the wild type clamp included and excluded. The authors determined that without the wild type clamp, they could not achieve sensitivity at the desired levels.
They then evaluated the sensitivity of their clamp assay by looking at the electrochemical signal relative to varying concentrations of RNA in the solution. They found a limit of detection of 1 fg μl-1 and a turnaround time of five minutes. Finally, to demonstrate that this clamp assay was applicable to other mutations, they tested for biomarkers in the BRAF gene. The BRAF studies worked similarly to the KRAS studies, demonstrating the general applicability of this technique.
After establishing the integrity of their clamp assay system, Das et al. then tested whether their system could detect cell free nucleic acids from serum retrieved from patients with lung cancer and patients with melanoma. These samples were compared to healthy donors and validated using an established PCR clamp method. Importantly, they tested both purified samples and samples taken directly from the patients without any purification. Their clamp assay identified 3/14 lung cancer patients as positive for KRAS and 4/9 melanoma patients as positive for BRAF in both the purified and non-purified samples.
This work illustrates a new method for detecting cell free nucleic acids using peptide nucleic acid clamps and a nanostructured microelectrode chip. It is sufficiently sensitive and selective to detect cancer biomarkers in patient blood. Compared to other methods it is cost effective, minimally invasive, and requires little sample preparation.
More information: "An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum" Nature Chemistry, (2015) DOI: 10.1038/nchem.2270
Abstract
The analysis of cell-free nucleic acids (cfNAs), which are present at significant levels in the blood of cancer patients, can reveal the mutational spectrum of a tumour without the need for invasive sampling of the tissue. However, this requires differentiation between the nucleic acids that originate from healthy cells and the mutated sequences shed by tumour cells. Here we report an electrochemical clamp assay that directly detects mutated sequences in patient serum. This is the first successful detection of cfNAs without the need for enzymatic amplification, a step that normally requires extensive sample processing and is prone to interference. The new chip-based assay reads out the presence of mutations within 15 minutes using a collection of oligonucleotides that sequester closely related sequences in solution, and thus allow only the mutated sequence to bind to a chip-based sensor. We demonstrate excellent levels of sensitivity and specificity and show that the clamp assay accurately detects mutated sequences in a collection of samples taken from lung cancer and melanoma patients.
Journal information: Nature Chemistry
© 2015 Phys.org