Bio researchers receive patent to fight superbugs

Superbugs, antibiotic-resistant bacteria, have been on the rise since antibiotics were first introduced 80 years ago. That's because these germ-fighting agents have lost their punch from being overprescribed and misused, allowing bacteria pathogens to develop immunities against them.
As a result, superbugs sicken nearly 2 million Americans each year and roughly 23,000 people die annually from these infections, according to the Centers for Disease Control and Prevention. Many more people die from other conditions that are complicated by antibiotic-resistant infections.
Lawrence Livermore National Laboratory scientists have figured out a way to reverse this trend by developing novel antibiotics that effectively kill superbugs. They were recently issued a patent for producing antimicrobial compounds that degrade and destroy antibiotic-resistant bacteria by using the pathogen's own genes against it. Their approach can be used to fight superbugs such as antibiotic-resistant E. coli, Salmonella, Campylobacter, Methicillin-resistant Staphylococcus aureus (MRSA), Bacillus anthracis and many others.
Antimicrobial compounds available today are designed to treat a host of bacteria, not targeting specific ones. While this is effective in the short term, bacteria are able to modify themselves over time to prevent a number of antibiotics from inhibiting their growth, rendering the antibiotics useless. To prevent this, the LLNL team created a process to discover new antibiotics designed to eradicate targeted bacterium responsible for a specific infection.
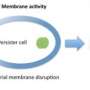
Their technique uses computational tools and genome sequencing to identify which genes inside a bacterium encode for lytic proteins that can be used to kill it. Lytic proteins are enzymes that normally produce nicks in cell walls that allow cells to divide and multiply. But used in high concentrations, these enzymes cause rapid cell-wall degradation and cell rupture, a process known as lysis. Lytic proteins circumvent any defenses that bacteria have developed against today's broadly focused antimicrobials.
Once genes encoding lytic proteins are identified in a bacterium, Lab researchers synthesize those genes and produce the lytic proteins in a laboratory setting. They are then experimentally introduced to the cell walls of targeted pathogens to destroy them.
"We knew about these proteins because they were primarily coded and identified in bacteriophages, virus particles that attack bacterium and make copies of themselves," said Matthew Coleman, a member of the LLNL research team. "Bacteriophages need a way to get out of the bacteria once they make enough copies. So they encode a lytic protein, called an endolysin, that breaks down the cell wall, causing the cell to die. The wall is what gives the cell its strength."
The team's unique discovery was the existence of lytic proteins that are not associated with bacteriophages. They learned that bacteria themselves encode their own versions of these lytic proteins, which they use under tight regulatory control to remodel their cell wall during cell division. Tight regulatory productions means that lytic protein is not overproduced because too much of it will kill the cells. The team conducted lab experiments that show lytic protein produced by bacteria killed pathogens more effectively than lytic protein produced by bacteriophages.
LLNL's resources allowed the team to sequence genomes within a few weeks to obtain information on new pathogens, unknown bacteria species and even species yet to be sequenced. This data helps the team determine which genes encode lytic proteins and reproduce those proteins in large quantities to eliminate targeted pathogens.
So far, the team's novel approach has been extremely effective in lab testing. In CDC experiments, the LLNL-produced lytic protein killed 100 percent of Bacillus anthracis cells, the bacterium that causes anthrax.
"We can isolate these genes to support the development of effective antimicrobial agents for commercial and national needs," Coleman said.
The team hopes to continue their experiments with different types of antibiotic-resistant bacteria. Its members also include LLNL researchers Feliza Bourguet, Paul Jackson, Brian Souza and Patrik D'haeseleer.
Provided by Lawrence Livermore National Laboratory