Antibiotic resistance enzyme caught in the act
Resistance to an entire class of antibiotics – aminoglycosides—has the potential to spread to many types of bacteria, according to new biochemistry research.
A mobile gene called NpmA was discovered in E. coli bacteria isolated from a Japanese patient several years ago. Global spread of NpmA and related antibiotic resistance enzymes could disable an entire class of tools doctors use to fight serious or life-threatening infections.
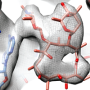
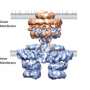
Using X-ray crystallography, researchers at Emory University School of Medicine made an atomic-scale snapshot of how the enzyme encoded by NpmA interacts with part of the ribosome, protein factories essential for all cells to function. NpmA imparts a tiny chemical change that makes the ribosome, and the bacteria, resistant to the drugs' effects.
The results, published in PNAS Early Edition, provide clues to the threat NpmA poses, but also reveal potential targets to develop drugs that could overcome resistance from this group of enzymes.
First author of the paper is postdoctoral fellow Jack Dunkle, PhD. Co-senior authors are assistant professor of biochemistry Christine Dunham, PhD and associate professor of biochemistry Graeme Conn, PhD.
Doctors generally use aminoglycoside antibiotics only for serious infections, because they can be toxic to the kidney and inner ear. But the growing problem of resistance to other types of antibiotics has sparked renewed interest in aminoglycosides' clinical use.
Examples of aminoglycosides include: streptomycin (the first antibiotic remedy for tuberculosis), kanamycin, tobramycin (often used in cystic fibrosis), gentamicin and neomycin.
Aminoglycosides bind to ribosomes, interfering with protein production in bacteria. Most mobile genes that confer aminoglycoside resistance chemically alter the antibiotics, and are active against only a few antibiotics. Instead, the NpmA-encoded enzyme modifies the ribosome so that aminoglycoside antibiotics don't interfere with it anymore. That's why it's more dangerous.
Another feature of NpmA that makes it dangerous is that it recognizes structural features that are common to all bacterial ribosomes. The information the Emory team obtained suggests that NpmA, found in E. coli, could easily work in other types of bacteria.
The structures of ribosome alone and the NpmA enzyme alone were already available; the Emory team was able to capture the two together in a "pre-catalytic state."
Aminoglycosides are naturally produced by certain types of soil bacteria against other bacteria, and the producer bacteria have to make a resistance enzyme to prevent self-poisoning. Scientists hypothesize that the genes that encode this type of resistance enzymes in pathogenic bacteria were originally acquired from an aminoglycoside producer, Conn says.
More information: Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistant methyltransferase NpmA, PNAS, www.pnas.org/cgi/doi/10.1073/pnas.1402789111
Journal information: Proceedings of the National Academy of Sciences
Provided by Emory University