Better chemistry through parallel in time algorithms
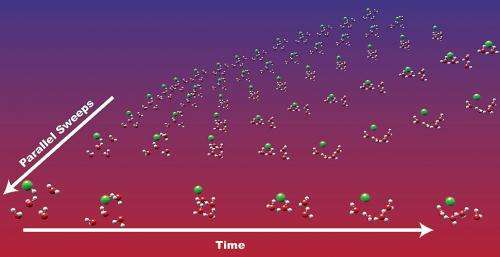
Molecular dynamics simulations often take too long to be practical for simulating chemical processes that occur on long timescales. Scientists DOE's Pacific Northwest National Laboratory, the University of Chicago, and the University of California at San Diego showed that time integration algorithms working in parallel can significantly speed up computationally demanding molecular dynamics simulations, opening new avenues for studying complex, long-lasting processes as diverse as carbon sequestration and energy production and storage.
Molecular dynamics simulations provide valuable data about the physical movements and interactions of atoms and molecules over time. Unlike classical approaches, ab initio molecular dynamics (AIMD) simulations accurately calculate the movements of electrons, enabling scientists to study chemical reactions that involve breaking or forming covalent bonds. Although AIMD simulations are useful in areas such as industrial and biological catalysis, their use is limited because they are computationally costly.
More time-efficient simulations have been made possible by massively parallel supercomputers and parallel algorithms, which distribute the computational workload across different processors or computers. Still, AIMD approaches currently take several months to simulate events that span picoseconds, even though many important chemical processes take much longer.
To address this problem, the researchers tested several parallel algorithms that distribute computations for different time intervals of a chemical event to different processors. These parallel in time algorithms sped up a conventional molecular dynamics simulation of 1,000 silicon atoms by a factor of 3. They also sped up a challenging AIMD simulation of an atmospherically important chemical reaction involving hydrochloric acid by a factor of 14. When the AIMD simulation of this chemical process was done on the massively parallel supercomputer, the use of parallel in time algorithms compared with sequential algorithms reduced the duration of each computational time step from 32 to 7 seconds. Computing resources were used at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE's Office of Biological and Environmental Research, and the National Energy Research Scientific Computing Center.
The parallel in time algorithms are suitable for cloud computing. The speedup provided by these algorithms occurred even when they were implemented on machines connected by very slow networks. Moreover, these algorithms can be implemented using scripting languages, as well as standard quantum chemistry packages. Taken together, the findings demonstrate that parallel in time algorithms can allow researchers to use powerful AIMD simulations to study realistic and complex chemical processes that take place on long timescales.
More information: Bylaska EJ, JQ Weare, and JH Weare. 2013. "Extending molecular simulation time scales: Parallel in time integrations for high-level quantum chemistry and complex force representations." Journal of Chemical Physics 139(7):074114. DOI: 10.1063/1.4818328.
Journal information: Journal of Chemical Physics
Provided by US Department of Energy