Advanced computer simulations reveal the conformational changes of an enzyme anchoring to its substrate
Pathogenic bacteria bristle with surface proteins that help them to infect their host. From chemical hooks that anchor the bacteria in place to cloaking proteins that hide them from their host's immune system, each protein is clipped into place by an enzyme called sortase. Kei Moritsugu and his colleagues at the RIKEN Research Cluster for Innovation in Wako have now shown, in atomic detail, how this enzyme functions. The work could lead to new antibiotics that work by disrupting these functions.
Sortase is a difficult enzyme to study because it incorporates a highly flexible section within its structure known as an intrinsically disordered region (IDR). Previous research showed that the enzyme performs its protein-tethering function with the help of a calcium ion, but the molecular mechanism by which calcium interacts with the IDR to assist this process remained unclear.
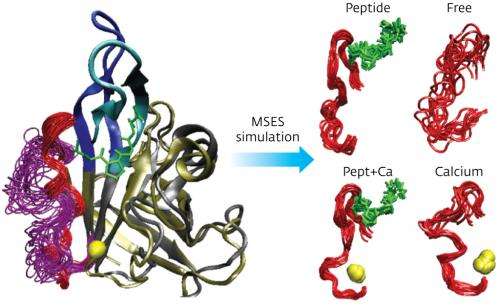
To find answers, Moritsugu and his team developed a computational simulation technique that they called multi-scale enhanced sampling (MSES). Conventional conformational sampling methods at the atomic scale are only applicable to the smallest of enzymes, so are not suitable for studying sortase. "The MSES simulation has overcome this problem," Moritsugu explains. The new simulation technique uses a multi-scale approach: it couples a detailed 'all-atom' structure with a coarse-grained model of the enzyme in a way that enhances sampling at the main point of interest, the IDR.
Using this approach, the researchers could calculate how sortase's structure changes as the protein substrate and the calcium ion bind to it. Their results showed that each binding partner induces a disorder-to-order transition in one part of the IDR, so that with both partners in place the entire IDR becomes ordered. The simulation also showed that with the calcium ion bound, the enzyme's structure shifts to better bind the protein substrate, explaining calcium's beneficial effect.
"Our discovery has medical implications," says Moritsugu, because blocking the enzyme's function could offer a new way to treat drug-resistant pathogens such as Methicillin-resistant Staphylococcus aureus(MRSA). "Sortase enzymes play a crucial role in virulence, infection, and colonization by pathogens. Elucidating the sortase function at atomistic resolution is expected to allow the computational design of inhibitors," he says.
The team is planning to use the MSES simulation technique for other biomolecules that have proven too large for conventional simulation techniques. Using the K computer, a powerful new supercomputer which has been jointly developed by RIKEN and Fujitsu, even multi-enzyme complexes could be studied using the technique, says Moritsugu.
More information: Moritsugu, K., Terada, T. & Kidera, A. Disorder-to-order transition of an intrinsically disordered region of sortase revealed by multiscale enhanced sampling. Journal of the American Chemical Society 134, 7094–7101 (2012). dx.doi.org/10.1021/ja3008402
Journal information: Journal of the American Chemical Society
Provided by RIKEN