Predatory bacterial crowdsourcing
(Phys.org)—Move forward. High-five your neighbor. Turn around. Repeat.
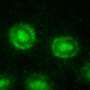
That's the winning formula of one of the world's smallest predators, the soil bacteria Myxococcus xanthus, and a new study by scientists at Rice University and the University of Texas Health Science Center at Houston (UTHealth) Medical School shows how M. xanthus uses the formula to spread, engulf and devour other bacteria.
The study, featured on the cover of this month's online issue of the journal PLOS Computational Biology, shows how the simple motions of individual bacteria are amplified within colonies of M. xanthus to form millions-strong waves moving outward in unison. The findings answer longstanding questions about how the waves form and the competitive edge they provide M. xanthus.
"When the cells at the edge of the colony are moving outward, they are unlikely to encounter another M. xanthus cell, so they keep moving forward," said lead author Oleg Igoshin, assistant professor of bioengineering at Rice. "When they are traveling the other way, back toward the rest of the colony, they are likely to encounter other cells of their kind, and when they pass beside one of these and touch, they get the signal to turn around."
Igoshin said the net effect is that the cells "spend more time moving outward than inward, and as a result, they spread faster."
M. xanthus is an oft-studied model organism in biology but, Igoshin said, it is one of the few well-studied organisms that lends itself to the study of systems biology, a rapidly growing field of life sciences that aims to model and discover emergent phenomena—like the rippling waves of M. xanthus colonies—that have a basis in genetics but only become apparent when cells cooperate.
"Most of the model bacteria that biologists selected for study in the lab were chosen because they were very good at growing on their own in a test tube and not sticking to the wall or to one another," Igoshin said. "When we were choosing model organisms, we lost a lot of the social properties that systems biologists like to study. M. xanthus is different in that people chose to study it because it grew into cool patterns and structures arising from cooperative behavior."
As a computational biologist, Igoshin specializes in creating mathematical models that accurately describe the behavior of living systems. Such models are useful for understanding the cellular and even genetic basis of emergent phenomena.
In the case of M. xanthus waves, Igoshin and Rice graduate student Haiyang Zhang and postdoctoral fellow Peng Shi created an agent-based model, a computer program that simulated the actions and interactions of individual cells to examine how they collectively produced M. xanthus waves.
The model showed that just three ingredients were needed to generate the rippling behavior:
- When two cells moving toward one another have side-to-side contact, they exchange a signal that causes one of them to reverse.
- A time interval after each reversal during which cells cannot reverse again.
- Physical interactions that cause the cells to align.
"We also found an interesting flip side for the behavior, which was counterintuitive and unexpected," Kaplan said. "The same behavior that causes the waves to spread quickly and to cover newly found prey also allows M. xanthus cells to stay on a patch of food and not drift away until the food is devoured."
The exact biochemical signals that the cells use to sense their prey and signal one another to reverse are still a mystery, but the new study helps to narrow the field. For example, many signals between bacteria do not require physical contact, but the study found that the reversing behavior requires contact between cells.
"If the mechanism for this behavior can be found, it could prove useful for synthetic biologists who are interested in programming touch-induced functions into synthetic organisms," Igoshin said.
More information: www.ploscompbiol.org/article/i … journal.pcbi.1002715
Journal information: PLoS Computational Biology
Provided by Rice University