Study reveals how protein machinery binds and wraps DNA to start replication
(PhysOrg.com) -- Before any cell - healthy or cancerous - can divide, it has to replicate its DNA. So scientists who want to know how normal cells work - and perhaps how to stop abnormal ones - are keen to understand this process. As a step toward that goal, scientists at the U.S. Department of Energy's Brookhaven National Laboratory and collaborators have deciphered molecular-level details of the complex choreography by which intricate cellular proteins recognize and bind to DNA to start the replication process. The study is published in the March 7, 2012, issue of the journal Structure.
"Every cell starts to replicate its genome at defined DNA sites called 'origins of replication,'" said Huilin Li, a biologist at Brookhaven Lab and Stony Brook University, who led the study. "A cell finds those origins in its vast genome with a protein 'machine' called the 'origin recognition complex,' or ORC."
In a typical bacterial genome, comprised of several million base pairs - the "letters" of the genetic code - there is only one such origin. However, in more complex eukaryotic organisms, such as humans with a genome of 3.4 billion base pairs, there may be tens of thousands of replication origins so that DNA replication can be carried out simultaneously at these sites to duplicate the genome in a reasonable time.
The goal of the current effort was to understand the first steps of the enormously complex task of duplicating a eukaryotic genome: how the protein machinery ORC recognizes and binds to the origin DNA, and how the origin-bound ORC enables the attachment of additional protein machinery that unwinds the DNA double helix into two single strands in preparation for DNA copying.
"This level of detail on the shape of the origin recognition complex and its interaction with DNA provides insight into a key cellular process, the initiation of DNA replication," said Daniel Janes, who oversees DNA replication grants at the National Institutes of Health's National Institute of General Medical Sciences, which partially supported the work. "Because DNA replication is closely tied to cell division, a thorough understanding of the process may lead to new ways to fight the uncontrolled cell division that characterizes cancer."
Previous studies have looked at similar but simpler protein machines in bacteria and other prokaryotes. In eukaryotic organisms, which have more complex cellular structure, the proteins themselves are more complicated - and larger - making them harder to study.
Some studies have looked at the eukaryotic proteins in relatively low resolution and in isolation. But none has taken a more detailed look and included how they bind with DNA - until now.
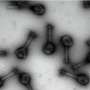
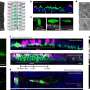
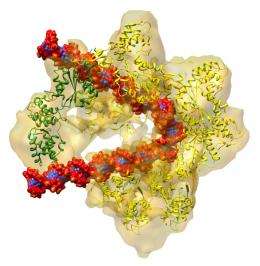
Jingchuan (Jim) Sun, a Brookhaven biologist who works with Li, used an imaging method known as cryo-electron microscopy to make higher resolution images of the eukaryotic ORC, in isolation, as it binds to DNA, and one step further in the process, when another protein unit binds to activate the entire structure. The research team used proteins from baker's yeast, which is a model system for eukaryotes.
The cryo-EM images produced a map, or outline, of the entire ORC structure as it changes during the activation process.
To explore the details of these changes, the scientists then turned to atomic-level x-ray crystal structures of small protein subunits that had been produced by other scientists. These subunit structures were made from prokaryotic cells known as archaea, which are evolutionarily related to eukaryotes, and so could serve as "stand-ins" for the eukaryotic subunit structures, which are still unknown.
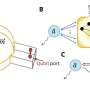
By fitting these subunits into the cryo-EM maps, the scientists were able to propose a detailed structure and mechanism for how the ORC may work: In simplest form, it starts as a two-lobed, crescent-shaped protein complex that wraps around and bends the origin DNA along the interior curve of the crescent. Sequential binding of a "replication initiator" known as Cdc6 (for cell division cycle 6) then induces a significant conformational change in the origin-bound ORC structure.
This structural conformation, the scientists say, is likely what opens the way for the attachment of the next piece of protein machinery essential to the DNA-replication process - the one that unwinds the two strands of the DNA double helix so that each can be copied.
"Our study is at a very basic level, trying to answer the fundamental biological questions about how this process works," said Li. "But our work has strong implications for health and disease, because unregulated or disregulated chromosomal duplication and uncontrolled cellular proliferation are the hallmarks of cancer. So understanding details of the mechanisms of DNA replication could potentially lead to new ways to fight cancer," he said.
Provided by Brookhaven National Laboratory