Deciphering distinct atomic motions in proteins with dynamic neutron scattering
Whether inside algae converting biomass to fuels or human cells responding to radiation exposure, proteins change their shape via atomic motions to perform a specific function. Today these shape-changing processes are still difficult to measure and understand. Scientists recently determined three classes of atomic motion using neutron scattering coupled with computational simulations.
Providing a viable approach to quantify specific types of atomic motion that can be linked to proteins' biological functions could enable a detailed understanding necessary for designing biobased or bio-inspired materials for energy production, energy storage, and other uses.
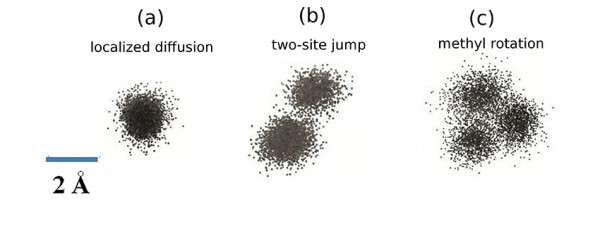
Flexibility or "softness" is required for proteins to carry out their catalytic and other biological activities. This flexibility conferred by the motion of atoms includes components from spring-like atomic vibrations, rotation of atoms about chemical bonds, random jumps, and diffusive motion. In this research, scientists obtained a comprehensive description of these components in a protein by combining dynamic neutron scattering experiments with molecular dynamics computer simulations to interpret the scattering data. Extracted data from this approach clearly show that each of these components is associated with a characteristic signal, in terms of length and time. The signature of elastic vibrations of isolated atoms within a single energy well was designated as localized diffusion and separated from true conformational changes, such as two-site jumps and methyl rotations which are between energy wells. The new approach of coupling advanced neutron scattering with high-performance computing could enable a better understanding of how proteins function and inform the design of protein-based biomaterials for various functions.
Publication details
"Three classes of motion in the dynamic neutron-scattering susceptibility of a globular protein," Physical Review Letters 107, 148102, 2011. DOI: 10.1103/PhyRevLett.107.148102
"Surface hydration amplifies single-well protein atom diffusion propagating into the macromolecular core," Physical Review Letters 108, 238102, 2012. DOI: 10.1103/PhyRevLett.108.238102
"Elastic and conformational softness of a globular protein," Physical Review Letters 110, 028104, 2013. DOI: 10.1103/PhyRevLett.110.028104
Journal information: Physical Review Letters
Provided by US Department of Energy