Key to pathogenic slime uncovered
(Phys.org) —Dental plaque, the sludge in hot springs and black slime inside of toilets are all examples of biofilms, made of slick communities of bacteria that also play roles in many diseases.
Now, a new study reveals the intricacies of how bacteria adhere to surfaces and form biofilms, a discovery that could lead to treatments for urinary tract infections and cystic fibrosis by countering antibiotic tolerance in bacteria.
The study, published Sept. 2 in the journal eLife, details how a ubiquitous bacterial signaling molecule called cyclic-di-GMP plays a key role in the switch from free-swimming cultures to biofilms.
"Most bacteria tend to form biofilms as an adaptation strategy to their changing environment," said Holger Sondermann, associate professor of molecular medicine at Cornell and one of the paper's senior authors. Environmental triggers prompt bacteria to secrete slime made of DNA, sugars and proteins that anchor bacterial colonies to surfaces.
The study reports on biofilm formation in the largely nonpathogenic bacteria Pseudomonas fluorescens, commonly found on plant roots.
Environmental cues such as the nutrient phosphate prompt P. fluorescens to form biofilms via sophisticated signaling systems that involve cyclic-di-GMP.
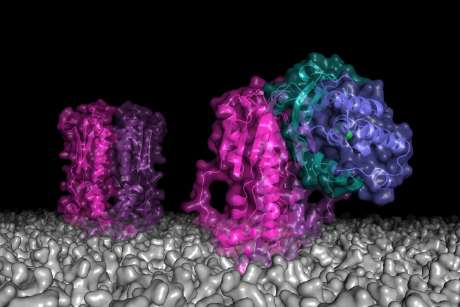
When P. fluorescens experiences nutrient-rich conditions, levels of cyclic-di-GMP in the cell rise, which is sensed by a protein called LapD. In turn, LapD engages LapG, an enzyme that in its free form is responsible for breaking apart another protein important for P. fluorescens' tight adhesion to surfaces.
In other words, when the LapG enzyme is bound, P. fluorescens can attach to a surface and a biofilm can form.
When phosphate levels are low, this cascade of events occurs in reverse, freeing the LapG enzyme to break the bonds that hold P. fluorescens cells to surfaces, which causes biofilms to disperse.
This system, which is also found in other bacteria, may be exploited as an Achilles' heel of biofilm formation. For example, the researchers showed that small molecules called peptides can mimic the docking sites for LapG on LapD, thereby preventing their binding and leaving LapG available to prevent biofilms from forming. Such findings could be used to develop drugs to disrupt the ability of bacteria to bind to a surface and impede biofilms.
"Some 80 percent of chronic infections have been attributed to biofilm formation," Sondermann said. They also clog up catheters, heart pacemakers and heart valves. Furthermore, biofilms may protect bacteria from antibiotics and contribute to antiobiotic resistance.
Journal information: eLife
Provided by Cornell University